
Puumala virus (strain Sotkamo/V-2969/81)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Hantaviridae; Mammantavirinae; Orthohantavirus; Puumala orthohantavirus
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
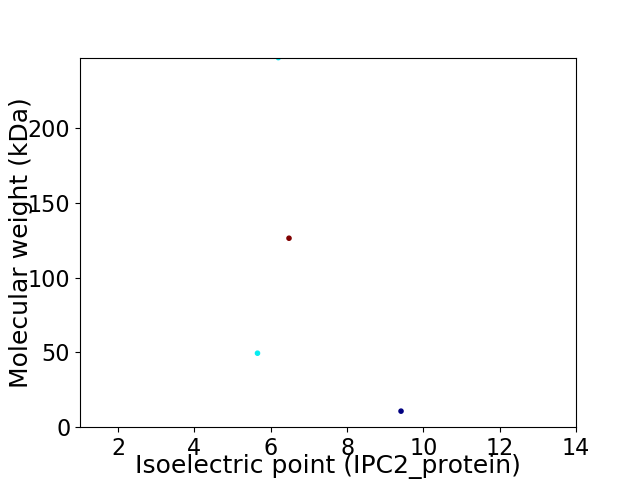
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
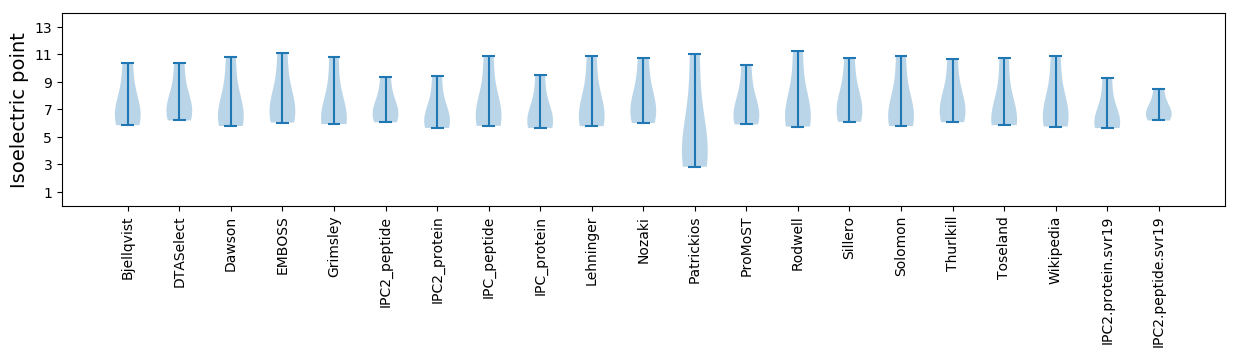
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P27313|NCAP_PUUMS Nucleoprotein OS=Puumala virus (strain Sotkamo/V-2969/81) OX=39002 GN=N PE=3 SV=1
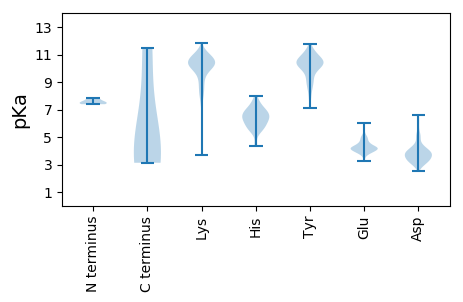
MM1 pKa = 7.84SDD3 pKa = 3.19LTDD6 pKa = 3.28IQEE9 pKa = 4.92DD10 pKa = 3.73ITRR13 pKa = 11.84HH14 pKa = 4.34EE15 pKa = 4.1QQLIVARR22 pKa = 11.84QKK24 pKa = 11.27LKK26 pKa = 10.1DD27 pKa = 3.35AEE29 pKa = 4.31RR30 pKa = 11.84AVEE33 pKa = 4.02VDD35 pKa = 3.33PDD37 pKa = 3.98DD38 pKa = 4.16VNKK41 pKa = 9.1NTLQARR47 pKa = 11.84QQTVSALEE55 pKa = 4.66DD56 pKa = 3.13KK57 pKa = 11.04LADD60 pKa = 3.77YY61 pKa = 9.96KK62 pKa = 11.1RR63 pKa = 11.84RR64 pKa = 11.84MADD67 pKa = 2.79AVSRR71 pKa = 11.84KK72 pKa = 10.22KK73 pKa = 10.07MDD75 pKa = 3.75TKK77 pKa = 10.32PTDD80 pKa = 3.46PTGIEE85 pKa = 4.1PDD87 pKa = 3.36DD88 pKa = 3.93HH89 pKa = 7.87LKK91 pKa = 10.48EE92 pKa = 4.18RR93 pKa = 11.84SSLRR97 pKa = 11.84YY98 pKa = 10.4GNVLDD103 pKa = 4.38VNAIDD108 pKa = 3.67IEE110 pKa = 4.45EE111 pKa = 4.59PSGQTADD118 pKa = 3.07WYY120 pKa = 9.87TIGVYY125 pKa = 10.42VIGFTLPIILKK136 pKa = 9.61ALYY139 pKa = 8.83MLSTRR144 pKa = 11.84GRR146 pKa = 11.84QTVKK150 pKa = 10.03EE151 pKa = 4.09NKK153 pKa = 7.61GTRR156 pKa = 11.84IRR158 pKa = 11.84FKK160 pKa = 10.94DD161 pKa = 3.35DD162 pKa = 2.88TSFEE166 pKa = 5.01DD167 pKa = 3.01INGIRR172 pKa = 11.84RR173 pKa = 11.84PKK175 pKa = 9.58HH176 pKa = 6.21LYY178 pKa = 10.21VSMPTAQSTMKK189 pKa = 10.72AEE191 pKa = 4.02EE192 pKa = 4.06LTPGRR197 pKa = 11.84FRR199 pKa = 11.84TIVCGLFPTQIQVRR213 pKa = 11.84NIMSPVMGVIGFSFFVKK230 pKa = 10.14DD231 pKa = 2.89WSEE234 pKa = 4.34RR235 pKa = 11.84IRR237 pKa = 11.84EE238 pKa = 4.09FMEE241 pKa = 4.24KK242 pKa = 9.51EE243 pKa = 4.07CPFIKK248 pKa = 10.5PEE250 pKa = 4.12VKK252 pKa = 10.17PGTPAQEE259 pKa = 3.72IEE261 pKa = 3.9MLKK264 pKa = 10.37RR265 pKa = 11.84NKK267 pKa = 9.79IYY269 pKa = 10.33FMQRR273 pKa = 11.84QDD275 pKa = 3.84VLDD278 pKa = 4.28KK279 pKa = 11.52NHH281 pKa = 6.16VADD284 pKa = 3.67IDD286 pKa = 3.88KK287 pKa = 11.09LIDD290 pKa = 3.44YY291 pKa = 7.82AASGDD296 pKa = 3.84PTSPDD301 pKa = 3.95NIDD304 pKa = 3.74SPNAPWVFACAPDD317 pKa = 3.86RR318 pKa = 11.84CPPTCIYY325 pKa = 9.3VAGMAEE331 pKa = 4.22LGAFFSILQDD341 pKa = 3.28MRR343 pKa = 11.84NTIMASKK350 pKa = 9.3TVGTAEE356 pKa = 4.09EE357 pKa = 4.16KK358 pKa = 10.68LKK360 pKa = 10.88KK361 pKa = 10.03KK362 pKa = 10.65SSFYY366 pKa = 10.41QSYY369 pKa = 10.61LRR371 pKa = 11.84RR372 pKa = 11.84TQSMGIQLDD381 pKa = 3.34QRR383 pKa = 11.84IILLFMLEE391 pKa = 4.08WGKK394 pKa = 11.05EE395 pKa = 4.19MVDD398 pKa = 3.77HH399 pKa = 6.53FHH401 pKa = 7.32LGDD404 pKa = 4.56DD405 pKa = 4.07MDD407 pKa = 4.87PEE409 pKa = 4.67LRR411 pKa = 11.84GLAQALIDD419 pKa = 3.78QKK421 pKa = 11.41VKK423 pKa = 10.55EE424 pKa = 4.47ISNQEE429 pKa = 3.84PLKK432 pKa = 10.76II433 pKa = 4.1
MM1 pKa = 7.84SDD3 pKa = 3.19LTDD6 pKa = 3.28IQEE9 pKa = 4.92DD10 pKa = 3.73ITRR13 pKa = 11.84HH14 pKa = 4.34EE15 pKa = 4.1QQLIVARR22 pKa = 11.84QKK24 pKa = 11.27LKK26 pKa = 10.1DD27 pKa = 3.35AEE29 pKa = 4.31RR30 pKa = 11.84AVEE33 pKa = 4.02VDD35 pKa = 3.33PDD37 pKa = 3.98DD38 pKa = 4.16VNKK41 pKa = 9.1NTLQARR47 pKa = 11.84QQTVSALEE55 pKa = 4.66DD56 pKa = 3.13KK57 pKa = 11.04LADD60 pKa = 3.77YY61 pKa = 9.96KK62 pKa = 11.1RR63 pKa = 11.84RR64 pKa = 11.84MADD67 pKa = 2.79AVSRR71 pKa = 11.84KK72 pKa = 10.22KK73 pKa = 10.07MDD75 pKa = 3.75TKK77 pKa = 10.32PTDD80 pKa = 3.46PTGIEE85 pKa = 4.1PDD87 pKa = 3.36DD88 pKa = 3.93HH89 pKa = 7.87LKK91 pKa = 10.48EE92 pKa = 4.18RR93 pKa = 11.84SSLRR97 pKa = 11.84YY98 pKa = 10.4GNVLDD103 pKa = 4.38VNAIDD108 pKa = 3.67IEE110 pKa = 4.45EE111 pKa = 4.59PSGQTADD118 pKa = 3.07WYY120 pKa = 9.87TIGVYY125 pKa = 10.42VIGFTLPIILKK136 pKa = 9.61ALYY139 pKa = 8.83MLSTRR144 pKa = 11.84GRR146 pKa = 11.84QTVKK150 pKa = 10.03EE151 pKa = 4.09NKK153 pKa = 7.61GTRR156 pKa = 11.84IRR158 pKa = 11.84FKK160 pKa = 10.94DD161 pKa = 3.35DD162 pKa = 2.88TSFEE166 pKa = 5.01DD167 pKa = 3.01INGIRR172 pKa = 11.84RR173 pKa = 11.84PKK175 pKa = 9.58HH176 pKa = 6.21LYY178 pKa = 10.21VSMPTAQSTMKK189 pKa = 10.72AEE191 pKa = 4.02EE192 pKa = 4.06LTPGRR197 pKa = 11.84FRR199 pKa = 11.84TIVCGLFPTQIQVRR213 pKa = 11.84NIMSPVMGVIGFSFFVKK230 pKa = 10.14DD231 pKa = 2.89WSEE234 pKa = 4.34RR235 pKa = 11.84IRR237 pKa = 11.84EE238 pKa = 4.09FMEE241 pKa = 4.24KK242 pKa = 9.51EE243 pKa = 4.07CPFIKK248 pKa = 10.5PEE250 pKa = 4.12VKK252 pKa = 10.17PGTPAQEE259 pKa = 3.72IEE261 pKa = 3.9MLKK264 pKa = 10.37RR265 pKa = 11.84NKK267 pKa = 9.79IYY269 pKa = 10.33FMQRR273 pKa = 11.84QDD275 pKa = 3.84VLDD278 pKa = 4.28KK279 pKa = 11.52NHH281 pKa = 6.16VADD284 pKa = 3.67IDD286 pKa = 3.88KK287 pKa = 11.09LIDD290 pKa = 3.44YY291 pKa = 7.82AASGDD296 pKa = 3.84PTSPDD301 pKa = 3.95NIDD304 pKa = 3.74SPNAPWVFACAPDD317 pKa = 3.86RR318 pKa = 11.84CPPTCIYY325 pKa = 9.3VAGMAEE331 pKa = 4.22LGAFFSILQDD341 pKa = 3.28MRR343 pKa = 11.84NTIMASKK350 pKa = 9.3TVGTAEE356 pKa = 4.09EE357 pKa = 4.16KK358 pKa = 10.68LKK360 pKa = 10.88KK361 pKa = 10.03KK362 pKa = 10.65SSFYY366 pKa = 10.41QSYY369 pKa = 10.61LRR371 pKa = 11.84RR372 pKa = 11.84TQSMGIQLDD381 pKa = 3.34QRR383 pKa = 11.84IILLFMLEE391 pKa = 4.08WGKK394 pKa = 11.05EE395 pKa = 4.19MVDD398 pKa = 3.77HH399 pKa = 6.53FHH401 pKa = 7.32LGDD404 pKa = 4.56DD405 pKa = 4.07MDD407 pKa = 4.87PEE409 pKa = 4.67LRR411 pKa = 11.84GLAQALIDD419 pKa = 3.78QKK421 pKa = 11.41VKK423 pKa = 10.55EE424 pKa = 4.47ISNQEE429 pKa = 3.84PLKK432 pKa = 10.76II433 pKa = 4.1
Molecular weight: 49.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P0C760|L_PUUMS RNA-directed RNA polymerase L OS=Puumala virus (strain Sotkamo/V-2969/81) OX=39002 GN=L PE=3 SV=1
MM1 pKa = 7.56NSNLLLPDD9 pKa = 3.41KK10 pKa = 10.1NLRR13 pKa = 11.84MQRR16 pKa = 11.84EE17 pKa = 4.07QWKK20 pKa = 6.72WTQMTLIKK28 pKa = 8.13THH30 pKa = 6.83CKK32 pKa = 9.75PGNKK36 pKa = 8.62QCQHH40 pKa = 5.5WRR42 pKa = 11.84TNSQTTRR49 pKa = 11.84EE50 pKa = 4.58GWQMLCPGKK59 pKa = 10.44KK60 pKa = 9.18WILNLLTRR68 pKa = 11.84LGLNLMTTSRR78 pKa = 11.84RR79 pKa = 11.84DD80 pKa = 3.2QALGMEE86 pKa = 4.59MSLMM90 pKa = 4.12
MM1 pKa = 7.56NSNLLLPDD9 pKa = 3.41KK10 pKa = 10.1NLRR13 pKa = 11.84MQRR16 pKa = 11.84EE17 pKa = 4.07QWKK20 pKa = 6.72WTQMTLIKK28 pKa = 8.13THH30 pKa = 6.83CKK32 pKa = 9.75PGNKK36 pKa = 8.62QCQHH40 pKa = 5.5WRR42 pKa = 11.84TNSQTTRR49 pKa = 11.84EE50 pKa = 4.58GWQMLCPGKK59 pKa = 10.44KK60 pKa = 9.18WILNLLTRR68 pKa = 11.84LGLNLMTTSRR78 pKa = 11.84RR79 pKa = 11.84DD80 pKa = 3.2QALGMEE86 pKa = 4.59MSLMM90 pKa = 4.12
Molecular weight: 10.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3827 |
90 |
2156 |
956.8 |
108.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.775 ± 0.408 | 2.718 ± 1.066 |
5.592 ± 0.716 | 5.932 ± 0.582 |
4.599 ± 0.588 | 5.749 ± 0.658 |
2.09 ± 0.154 | 6.245 ± 0.361 |
6.768 ± 0.363 | 10.295 ± 0.618 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.535 ± 0.893 | 3.893 ± 0.399 |
3.946 ± 0.417 | 4.625 ± 0.517 |
4.599 ± 0.878 | 6.898 ± 0.631 |
6.323 ± 1.102 | 6.48 ± 0.724 |
1.516 ± 0.306 | 3.423 ± 0.55 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
