
Candidatus Electronema sp. GS
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobulbaceae; Candidatus Electronema; unclassified Candidatus Electronema
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
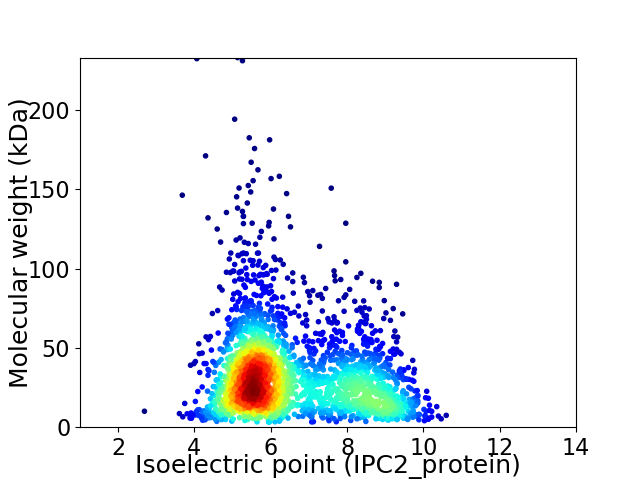
Virtual 2D-PAGE plot for 2614 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A521G4F0|A0A521G4F0_9DELT Diadenylate cyclase OS=Candidatus Electronema sp. GS OX=2005002 GN=dacA PE=3 SV=1
MM1 pKa = 7.05QNSVQTITVLTAALLLGATAASALPIKK28 pKa = 10.53SGNDD32 pKa = 2.94KK33 pKa = 10.98VQVQLYY39 pKa = 9.8GEE41 pKa = 4.11VDD43 pKa = 3.61RR44 pKa = 11.84AVMYY48 pKa = 10.96ADD50 pKa = 5.55DD51 pKa = 5.44GYY53 pKa = 11.09QDD55 pKa = 4.9KK56 pKa = 10.74IFHH59 pKa = 6.54VDD61 pKa = 3.1NTHH64 pKa = 5.68SEE66 pKa = 4.27TRR68 pKa = 11.84VGLNGEE74 pKa = 4.29VTATQCLTVGGNVEE88 pKa = 4.9LKK90 pKa = 10.14WEE92 pKa = 4.47ANPARR97 pKa = 11.84AVSMDD102 pKa = 3.53EE103 pKa = 3.92EE104 pKa = 4.82SISGEE109 pKa = 3.82FAAEE113 pKa = 4.24LIEE116 pKa = 4.52MYY118 pKa = 10.61INTANAGKK126 pKa = 10.4LSLGQGSTASDD137 pKa = 3.19ASSEE141 pKa = 4.06VDD143 pKa = 4.44LSGTDD148 pKa = 3.29IIGNAGVADD157 pKa = 4.51LGGGLSFYY165 pKa = 10.98DD166 pKa = 3.76AAVDD170 pKa = 4.84GYY172 pKa = 11.84SDD174 pKa = 3.76LTADD178 pKa = 3.7AVFHH182 pKa = 6.31GGISAEE188 pKa = 4.08GLGKK192 pKa = 10.07INRR195 pKa = 11.84VLYY198 pKa = 6.59EE199 pKa = 3.83TRR201 pKa = 11.84SFGGFTFGASAGEE214 pKa = 4.1EE215 pKa = 4.11EE216 pKa = 4.85VVDD219 pKa = 4.51FALSYY224 pKa = 11.0AGEE227 pKa = 4.2FNGNQLEE234 pKa = 4.69AKK236 pKa = 9.74AAWSNPGEE244 pKa = 4.75GYY246 pKa = 10.07NQINGSASLLLNSGLNFTVAASSKK270 pKa = 10.95DD271 pKa = 3.68VDD273 pKa = 3.57NMPAYY278 pKa = 10.41GDD280 pKa = 4.73DD281 pKa = 3.54PSFMYY286 pKa = 10.41GKK288 pKa = 9.72IGYY291 pKa = 8.9KK292 pKa = 9.83CDD294 pKa = 3.47QLSSFGSTAVSVDD307 pKa = 3.16YY308 pKa = 11.25GIFNNADD315 pKa = 3.67LLDD318 pKa = 4.5AEE320 pKa = 4.72QEE322 pKa = 3.93ATAIGVQFVQEE333 pKa = 4.02LSDD336 pKa = 3.94YY337 pKa = 9.68STQLYY342 pKa = 9.63AGYY345 pKa = 10.03RR346 pKa = 11.84AFSLEE351 pKa = 4.34DD352 pKa = 3.34NTGVDD357 pKa = 3.82YY358 pKa = 11.5EE359 pKa = 4.87DD360 pKa = 4.27ISVVMAGARR369 pKa = 11.84LSFF372 pKa = 4.08
MM1 pKa = 7.05QNSVQTITVLTAALLLGATAASALPIKK28 pKa = 10.53SGNDD32 pKa = 2.94KK33 pKa = 10.98VQVQLYY39 pKa = 9.8GEE41 pKa = 4.11VDD43 pKa = 3.61RR44 pKa = 11.84AVMYY48 pKa = 10.96ADD50 pKa = 5.55DD51 pKa = 5.44GYY53 pKa = 11.09QDD55 pKa = 4.9KK56 pKa = 10.74IFHH59 pKa = 6.54VDD61 pKa = 3.1NTHH64 pKa = 5.68SEE66 pKa = 4.27TRR68 pKa = 11.84VGLNGEE74 pKa = 4.29VTATQCLTVGGNVEE88 pKa = 4.9LKK90 pKa = 10.14WEE92 pKa = 4.47ANPARR97 pKa = 11.84AVSMDD102 pKa = 3.53EE103 pKa = 3.92EE104 pKa = 4.82SISGEE109 pKa = 3.82FAAEE113 pKa = 4.24LIEE116 pKa = 4.52MYY118 pKa = 10.61INTANAGKK126 pKa = 10.4LSLGQGSTASDD137 pKa = 3.19ASSEE141 pKa = 4.06VDD143 pKa = 4.44LSGTDD148 pKa = 3.29IIGNAGVADD157 pKa = 4.51LGGGLSFYY165 pKa = 10.98DD166 pKa = 3.76AAVDD170 pKa = 4.84GYY172 pKa = 11.84SDD174 pKa = 3.76LTADD178 pKa = 3.7AVFHH182 pKa = 6.31GGISAEE188 pKa = 4.08GLGKK192 pKa = 10.07INRR195 pKa = 11.84VLYY198 pKa = 6.59EE199 pKa = 3.83TRR201 pKa = 11.84SFGGFTFGASAGEE214 pKa = 4.1EE215 pKa = 4.11EE216 pKa = 4.85VVDD219 pKa = 4.51FALSYY224 pKa = 11.0AGEE227 pKa = 4.2FNGNQLEE234 pKa = 4.69AKK236 pKa = 9.74AAWSNPGEE244 pKa = 4.75GYY246 pKa = 10.07NQINGSASLLLNSGLNFTVAASSKK270 pKa = 10.95DD271 pKa = 3.68VDD273 pKa = 3.57NMPAYY278 pKa = 10.41GDD280 pKa = 4.73DD281 pKa = 3.54PSFMYY286 pKa = 10.41GKK288 pKa = 9.72IGYY291 pKa = 8.9KK292 pKa = 9.83CDD294 pKa = 3.47QLSSFGSTAVSVDD307 pKa = 3.16YY308 pKa = 11.25GIFNNADD315 pKa = 3.67LLDD318 pKa = 4.5AEE320 pKa = 4.72QEE322 pKa = 3.93ATAIGVQFVQEE333 pKa = 4.02LSDD336 pKa = 3.94YY337 pKa = 9.68STQLYY342 pKa = 9.63AGYY345 pKa = 10.03RR346 pKa = 11.84AFSLEE351 pKa = 4.34DD352 pKa = 3.34NTGVDD357 pKa = 3.82YY358 pKa = 11.5EE359 pKa = 4.87DD360 pKa = 4.27ISVVMAGARR369 pKa = 11.84LSFF372 pKa = 4.08
Molecular weight: 39.07 kDa
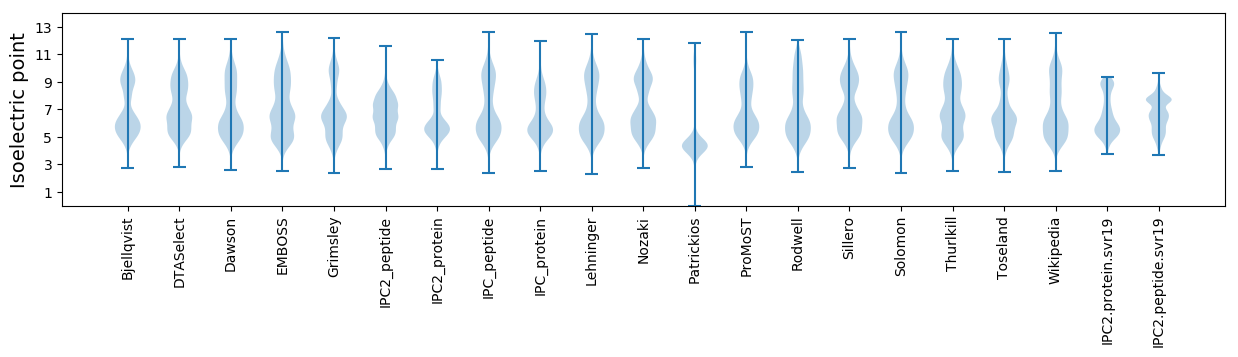
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A521G0Z2|A0A521G0Z2_9DELT Uncharacterized protein OS=Candidatus Electronema sp. GS OX=2005002 GN=CDV28_1196 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.48NIDD5 pKa = 3.65RR6 pKa = 11.84RR7 pKa = 11.84GVLPDD12 pKa = 3.34GLRR15 pKa = 11.84LPRR18 pKa = 11.84CPRR21 pKa = 11.84QSAKK25 pKa = 10.21IGCTRR30 pKa = 11.84VQRR33 pKa = 11.84WILLSCNGGYY43 pKa = 10.06FEE45 pKa = 4.66STTDD49 pKa = 3.02RR50 pKa = 11.84FTRR53 pKa = 11.84VQWLLSLSCKK63 pKa = 10.02LRR65 pKa = 11.84ADD67 pKa = 4.06YY68 pKa = 9.61PCRR71 pKa = 11.84AARR74 pKa = 11.84SGVRR78 pKa = 11.84PNAPAPSRR86 pKa = 11.84HH87 pKa = 5.96CRR89 pKa = 11.84GDD91 pKa = 3.13PCDD94 pKa = 4.56RR95 pKa = 11.84PMCRR99 pKa = 11.84HH100 pKa = 5.92IATMRR105 pKa = 11.84GEE107 pKa = 4.29YY108 pKa = 9.94KK109 pKa = 9.78IRR111 pKa = 11.84PYY113 pKa = 10.39RR114 pKa = 11.84PCRR117 pKa = 11.84AACNCAKK124 pKa = 10.4PLRR127 pKa = 11.84HH128 pKa = 5.65VSKK131 pKa = 10.11EE132 pKa = 3.91GWPMTSIICTAWLTCSSAGARR153 pKa = 11.84LHH155 pKa = 6.84RR156 pKa = 11.84SSS158 pKa = 3.93
MM1 pKa = 7.48KK2 pKa = 10.48NIDD5 pKa = 3.65RR6 pKa = 11.84RR7 pKa = 11.84GVLPDD12 pKa = 3.34GLRR15 pKa = 11.84LPRR18 pKa = 11.84CPRR21 pKa = 11.84QSAKK25 pKa = 10.21IGCTRR30 pKa = 11.84VQRR33 pKa = 11.84WILLSCNGGYY43 pKa = 10.06FEE45 pKa = 4.66STTDD49 pKa = 3.02RR50 pKa = 11.84FTRR53 pKa = 11.84VQWLLSLSCKK63 pKa = 10.02LRR65 pKa = 11.84ADD67 pKa = 4.06YY68 pKa = 9.61PCRR71 pKa = 11.84AARR74 pKa = 11.84SGVRR78 pKa = 11.84PNAPAPSRR86 pKa = 11.84HH87 pKa = 5.96CRR89 pKa = 11.84GDD91 pKa = 3.13PCDD94 pKa = 4.56RR95 pKa = 11.84PMCRR99 pKa = 11.84HH100 pKa = 5.92IATMRR105 pKa = 11.84GEE107 pKa = 4.29YY108 pKa = 9.94KK109 pKa = 9.78IRR111 pKa = 11.84PYY113 pKa = 10.39RR114 pKa = 11.84PCRR117 pKa = 11.84AACNCAKK124 pKa = 10.4PLRR127 pKa = 11.84HH128 pKa = 5.65VSKK131 pKa = 10.11EE132 pKa = 3.91GWPMTSIICTAWLTCSSAGARR153 pKa = 11.84LHH155 pKa = 6.84RR156 pKa = 11.84SSS158 pKa = 3.93
Molecular weight: 17.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
805550 |
28 |
2264 |
308.2 |
34.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.478 ± 0.055 | 1.432 ± 0.022 |
5.074 ± 0.034 | 6.76 ± 0.055 |
4.1 ± 0.039 | 7.163 ± 0.043 |
2.125 ± 0.022 | 6.141 ± 0.043 |
5.398 ± 0.052 | 10.627 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.444 ± 0.021 | 3.553 ± 0.031 |
4.326 ± 0.031 | 4.24 ± 0.042 |
5.656 ± 0.042 | 6.042 ± 0.044 |
4.964 ± 0.043 | 6.592 ± 0.036 |
1.144 ± 0.02 | 2.744 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
