
Bombyx mori cytoplasmic polyhedrosis virus (BmCPV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Cypovirus; Cypovirus 1
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
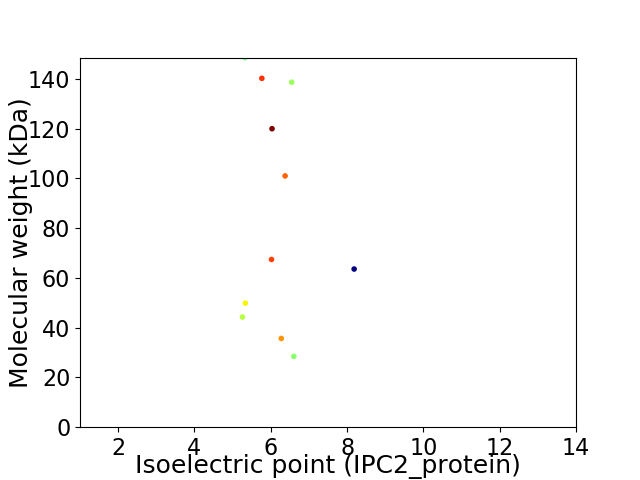
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7EWL8|C7EWL8_CPVBM p101 OS=Bombyx mori cytoplasmic polyhedrosis virus OX=110829 PE=4 SV=1
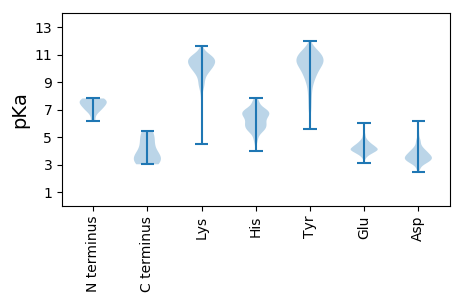
MM1 pKa = 6.17TTKK4 pKa = 10.63LFKK7 pKa = 9.87QTIIPTNKK15 pKa = 10.21DD16 pKa = 2.58VDD18 pKa = 3.49EE19 pKa = 4.94KK20 pKa = 11.12YY21 pKa = 10.44IYY23 pKa = 10.45FIEE26 pKa = 4.53RR27 pKa = 11.84ADD29 pKa = 3.61HH30 pKa = 6.96KK31 pKa = 10.38IIKK34 pKa = 8.48PLHH37 pKa = 6.06PAFEE41 pKa = 4.43PWQFAGNQHH50 pKa = 5.55AHH52 pKa = 5.34QLEE55 pKa = 3.91QGYY58 pKa = 11.15ADD60 pKa = 3.18GHH62 pKa = 5.75RR63 pKa = 11.84FITNYY68 pKa = 10.55NATSRR73 pKa = 11.84LNSAMTLQIMNKK85 pKa = 9.51PMITIVKK92 pKa = 9.74KK93 pKa = 8.79FTTNTPHH100 pKa = 6.4QVTTIPEE107 pKa = 4.8GYY109 pKa = 9.63SFGVVKK115 pKa = 10.68EE116 pKa = 4.08KK117 pKa = 11.13LCTNEE122 pKa = 3.78QLLSLLALTAEE133 pKa = 4.29PVIEE137 pKa = 4.67NDD139 pKa = 3.93GEE141 pKa = 4.45VNDD144 pKa = 5.21TIQEE148 pKa = 4.13DD149 pKa = 3.83EE150 pKa = 4.4TEE152 pKa = 4.16KK153 pKa = 11.2KK154 pKa = 9.42EE155 pKa = 4.49VEE157 pKa = 3.95MRR159 pKa = 11.84MNWSDD164 pKa = 3.84EE165 pKa = 4.1VVEE168 pKa = 4.62LPKK171 pKa = 10.46QQEE174 pKa = 4.06EE175 pKa = 4.62NVFVLSKK182 pKa = 10.5PSMISEE188 pKa = 4.29EE189 pKa = 4.16EE190 pKa = 4.03LMPIDD195 pKa = 4.33MEE197 pKa = 4.49VMTPKK202 pKa = 10.31VLEE205 pKa = 4.68PPTPLPSPIVVAVSSEE221 pKa = 4.13SPQVKK226 pKa = 9.91EE227 pKa = 4.49IEE229 pKa = 4.42RR230 pKa = 11.84LPADD234 pKa = 4.26LPQEE238 pKa = 4.01QRR240 pKa = 11.84VSRR243 pKa = 11.84KK244 pKa = 8.93PVARR248 pKa = 11.84TTKK251 pKa = 9.8PSFSSFSSPDD261 pKa = 2.97IVSANVQTPVPKK273 pKa = 9.97VAPRR277 pKa = 11.84PVEE280 pKa = 3.94EE281 pKa = 4.35FLSSVFTSLFEE292 pKa = 3.92AWFVRR297 pKa = 11.84VLPLIKK303 pKa = 9.9QGKK306 pKa = 6.86PVYY309 pKa = 9.73MMPGTIKK316 pKa = 10.19WEE318 pKa = 4.08RR319 pKa = 11.84EE320 pKa = 3.79AQITDD325 pKa = 3.01TKK327 pKa = 10.21TRR329 pKa = 11.84YY330 pKa = 9.34FNSVSGKK337 pKa = 10.14LILVEE342 pKa = 4.14MEE344 pKa = 4.35SITRR348 pKa = 11.84ALQPGEE354 pKa = 4.0LNDD357 pKa = 3.47RR358 pKa = 11.84RR359 pKa = 11.84EE360 pKa = 4.77LIRR363 pKa = 11.84QTLEE367 pKa = 4.13TVCAGHH373 pKa = 6.79NIHH376 pKa = 6.6VLTGKK381 pKa = 8.76DD382 pKa = 3.05TFGVLEE388 pKa = 4.36VEE390 pKa = 4.51
MM1 pKa = 6.17TTKK4 pKa = 10.63LFKK7 pKa = 9.87QTIIPTNKK15 pKa = 10.21DD16 pKa = 2.58VDD18 pKa = 3.49EE19 pKa = 4.94KK20 pKa = 11.12YY21 pKa = 10.44IYY23 pKa = 10.45FIEE26 pKa = 4.53RR27 pKa = 11.84ADD29 pKa = 3.61HH30 pKa = 6.96KK31 pKa = 10.38IIKK34 pKa = 8.48PLHH37 pKa = 6.06PAFEE41 pKa = 4.43PWQFAGNQHH50 pKa = 5.55AHH52 pKa = 5.34QLEE55 pKa = 3.91QGYY58 pKa = 11.15ADD60 pKa = 3.18GHH62 pKa = 5.75RR63 pKa = 11.84FITNYY68 pKa = 10.55NATSRR73 pKa = 11.84LNSAMTLQIMNKK85 pKa = 9.51PMITIVKK92 pKa = 9.74KK93 pKa = 8.79FTTNTPHH100 pKa = 6.4QVTTIPEE107 pKa = 4.8GYY109 pKa = 9.63SFGVVKK115 pKa = 10.68EE116 pKa = 4.08KK117 pKa = 11.13LCTNEE122 pKa = 3.78QLLSLLALTAEE133 pKa = 4.29PVIEE137 pKa = 4.67NDD139 pKa = 3.93GEE141 pKa = 4.45VNDD144 pKa = 5.21TIQEE148 pKa = 4.13DD149 pKa = 3.83EE150 pKa = 4.4TEE152 pKa = 4.16KK153 pKa = 11.2KK154 pKa = 9.42EE155 pKa = 4.49VEE157 pKa = 3.95MRR159 pKa = 11.84MNWSDD164 pKa = 3.84EE165 pKa = 4.1VVEE168 pKa = 4.62LPKK171 pKa = 10.46QQEE174 pKa = 4.06EE175 pKa = 4.62NVFVLSKK182 pKa = 10.5PSMISEE188 pKa = 4.29EE189 pKa = 4.16EE190 pKa = 4.03LMPIDD195 pKa = 4.33MEE197 pKa = 4.49VMTPKK202 pKa = 10.31VLEE205 pKa = 4.68PPTPLPSPIVVAVSSEE221 pKa = 4.13SPQVKK226 pKa = 9.91EE227 pKa = 4.49IEE229 pKa = 4.42RR230 pKa = 11.84LPADD234 pKa = 4.26LPQEE238 pKa = 4.01QRR240 pKa = 11.84VSRR243 pKa = 11.84KK244 pKa = 8.93PVARR248 pKa = 11.84TTKK251 pKa = 9.8PSFSSFSSPDD261 pKa = 2.97IVSANVQTPVPKK273 pKa = 9.97VAPRR277 pKa = 11.84PVEE280 pKa = 3.94EE281 pKa = 4.35FLSSVFTSLFEE292 pKa = 3.92AWFVRR297 pKa = 11.84VLPLIKK303 pKa = 9.9QGKK306 pKa = 6.86PVYY309 pKa = 9.73MMPGTIKK316 pKa = 10.19WEE318 pKa = 4.08RR319 pKa = 11.84EE320 pKa = 3.79AQITDD325 pKa = 3.01TKK327 pKa = 10.21TRR329 pKa = 11.84YY330 pKa = 9.34FNSVSGKK337 pKa = 10.14LILVEE342 pKa = 4.14MEE344 pKa = 4.35SITRR348 pKa = 11.84ALQPGEE354 pKa = 4.0LNDD357 pKa = 3.47RR358 pKa = 11.84RR359 pKa = 11.84EE360 pKa = 4.77LIRR363 pKa = 11.84QTLEE367 pKa = 4.13TVCAGHH373 pKa = 6.79NIHH376 pKa = 6.6VLTGKK381 pKa = 8.76DD382 pKa = 3.05TFGVLEE388 pKa = 4.36VEE390 pKa = 4.51
Molecular weight: 44.27 kDa
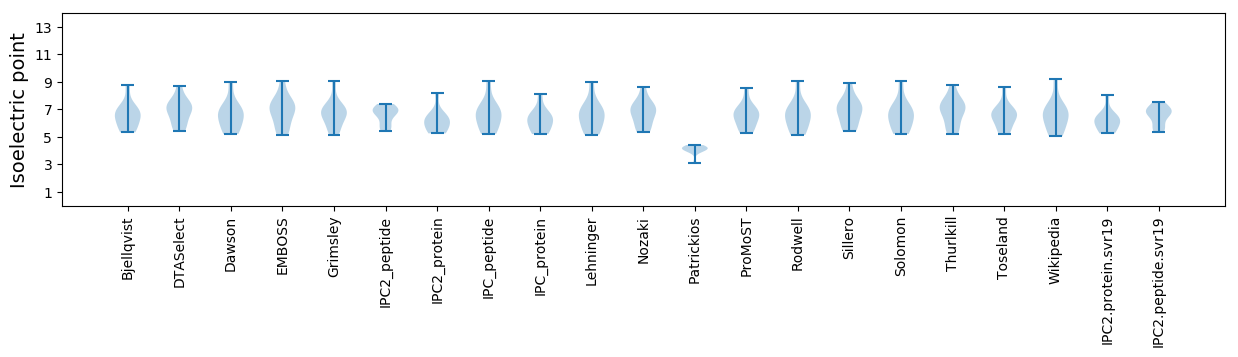
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D0EZK6|D0EZK6_CPVBM RNA-dependent RNA polymerase OS=Bombyx mori cytoplasmic polyhedrosis virus OX=110829 PE=1 SV=1
MM1 pKa = 7.39FAIDD5 pKa = 4.05PLKK8 pKa = 10.53HH9 pKa = 5.65SKK11 pKa = 10.57LYY13 pKa = 10.59EE14 pKa = 4.03EE15 pKa = 4.63YY16 pKa = 10.94GLYY19 pKa = 10.45LRR21 pKa = 11.84PHH23 pKa = 6.9QINQEE28 pKa = 3.99IKK30 pKa = 8.54PTTIKK35 pKa = 10.54KK36 pKa = 10.23KK37 pKa = 10.17EE38 pKa = 3.85LAPTIRR44 pKa = 11.84SIKK47 pKa = 8.95YY48 pKa = 9.15ASLIHH53 pKa = 7.22SMLAEE58 pKa = 3.8HH59 pKa = 7.52AARR62 pKa = 11.84HH63 pKa = 5.84NGTLINPRR71 pKa = 11.84MYY73 pKa = 11.37ADD75 pKa = 5.11MITLGNTKK83 pKa = 8.74VTVTKK88 pKa = 8.63GTPKK92 pKa = 10.7AQIDD96 pKa = 3.92TLKK99 pKa = 10.59MNGLTVVSKK108 pKa = 10.56SRR110 pKa = 11.84RR111 pKa = 11.84NNKK114 pKa = 9.1KK115 pKa = 10.34KK116 pKa = 10.17PVSDD120 pKa = 3.47TTATIDD126 pKa = 3.48EE127 pKa = 4.35NTNAIVTYY135 pKa = 10.29KK136 pKa = 10.83ALTEE140 pKa = 3.92MSTLIEE146 pKa = 4.27SFRR149 pKa = 11.84LPSGLTLIIFDD160 pKa = 3.97DD161 pKa = 4.06EE162 pKa = 5.1KK163 pKa = 10.96YY164 pKa = 10.59QSLIPNYY171 pKa = 8.93INQLIAYY178 pKa = 5.86TQPHH182 pKa = 7.71IIPTWQGIADD192 pKa = 4.32FSDD195 pKa = 3.56TYY197 pKa = 10.93LRR199 pKa = 11.84SYY201 pKa = 10.09FKK203 pKa = 10.89RR204 pKa = 11.84PFEE207 pKa = 4.06LTASNLAAPQKK218 pKa = 10.75HH219 pKa = 5.88NLSPMTRR226 pKa = 11.84SIFNNTGRR234 pKa = 11.84EE235 pKa = 3.82DD236 pKa = 3.4AVIRR240 pKa = 11.84KK241 pKa = 9.45LYY243 pKa = 10.72GYY245 pKa = 10.27GEE247 pKa = 4.44YY248 pKa = 11.12VFIKK252 pKa = 11.0YY253 pKa = 8.71EE254 pKa = 4.09GCLITWTGIYY264 pKa = 10.78GEE266 pKa = 4.2VTMMVNLSKK275 pKa = 10.66RR276 pKa = 11.84DD277 pKa = 3.55LGLDD281 pKa = 3.25VGDD284 pKa = 5.72DD285 pKa = 3.58YY286 pKa = 11.97LKK288 pKa = 10.45EE289 pKa = 4.1YY290 pKa = 10.77KK291 pKa = 10.13KK292 pKa = 10.91LLFYY296 pKa = 10.92GVITDD301 pKa = 5.76AIPSGISTRR310 pKa = 11.84STIMKK315 pKa = 9.91ISPHH319 pKa = 7.59KK320 pKa = 10.14MMNPSGGALAVLSKK334 pKa = 10.58FLEE337 pKa = 4.48AVVSTNVINATLVVYY352 pKa = 10.51AEE354 pKa = 4.34KK355 pKa = 10.85GAGKK359 pKa = 8.68TSFLSTYY366 pKa = 10.93AEE368 pKa = 4.08QLSLASGQVVGHH380 pKa = 6.99LSSDD384 pKa = 3.14AYY386 pKa = 10.61GRR388 pKa = 11.84WLAKK392 pKa = 9.89TKK394 pKa = 10.46DD395 pKa = 3.21IEE397 pKa = 4.52EE398 pKa = 4.17PSFAYY403 pKa = 10.15DD404 pKa = 3.46YY405 pKa = 11.4VLSLDD410 pKa = 3.57TDD412 pKa = 4.2DD413 pKa = 4.63NEE415 pKa = 4.92SYY417 pKa = 10.91YY418 pKa = 9.29EE419 pKa = 4.04QKK421 pKa = 10.93ASEE424 pKa = 4.65LLMSHH429 pKa = 7.16GISEE433 pKa = 4.36VAQYY437 pKa = 11.15EE438 pKa = 4.28LLSVRR443 pKa = 11.84KK444 pKa = 8.85KK445 pKa = 10.38IKK447 pKa = 10.4MMDD450 pKa = 3.36EE451 pKa = 4.0MNEE454 pKa = 3.76VLIAQLEE461 pKa = 4.3NADD464 pKa = 3.67THH466 pKa = 6.13SEE468 pKa = 4.04RR469 pKa = 11.84NFYY472 pKa = 11.58YY473 pKa = 9.9MVSTGKK479 pKa = 6.19TTPRR483 pKa = 11.84ILIVEE488 pKa = 4.06GHH490 pKa = 6.41FNAQDD495 pKa = 3.26ATIARR500 pKa = 11.84TDD502 pKa = 3.41TTVLLRR508 pKa = 11.84TINDD512 pKa = 3.35TTQAMRR518 pKa = 11.84DD519 pKa = 3.56RR520 pKa = 11.84QRR522 pKa = 11.84GGVVQLFLRR531 pKa = 11.84DD532 pKa = 3.14TYY534 pKa = 11.61YY535 pKa = 11.31RR536 pKa = 11.84LLPALHH542 pKa = 5.25TTVYY546 pKa = 8.96PFEE549 pKa = 4.09MLEE552 pKa = 4.54SIKK555 pKa = 10.15RR556 pKa = 11.84WKK558 pKa = 8.44WVHH561 pKa = 5.43
MM1 pKa = 7.39FAIDD5 pKa = 4.05PLKK8 pKa = 10.53HH9 pKa = 5.65SKK11 pKa = 10.57LYY13 pKa = 10.59EE14 pKa = 4.03EE15 pKa = 4.63YY16 pKa = 10.94GLYY19 pKa = 10.45LRR21 pKa = 11.84PHH23 pKa = 6.9QINQEE28 pKa = 3.99IKK30 pKa = 8.54PTTIKK35 pKa = 10.54KK36 pKa = 10.23KK37 pKa = 10.17EE38 pKa = 3.85LAPTIRR44 pKa = 11.84SIKK47 pKa = 8.95YY48 pKa = 9.15ASLIHH53 pKa = 7.22SMLAEE58 pKa = 3.8HH59 pKa = 7.52AARR62 pKa = 11.84HH63 pKa = 5.84NGTLINPRR71 pKa = 11.84MYY73 pKa = 11.37ADD75 pKa = 5.11MITLGNTKK83 pKa = 8.74VTVTKK88 pKa = 8.63GTPKK92 pKa = 10.7AQIDD96 pKa = 3.92TLKK99 pKa = 10.59MNGLTVVSKK108 pKa = 10.56SRR110 pKa = 11.84RR111 pKa = 11.84NNKK114 pKa = 9.1KK115 pKa = 10.34KK116 pKa = 10.17PVSDD120 pKa = 3.47TTATIDD126 pKa = 3.48EE127 pKa = 4.35NTNAIVTYY135 pKa = 10.29KK136 pKa = 10.83ALTEE140 pKa = 3.92MSTLIEE146 pKa = 4.27SFRR149 pKa = 11.84LPSGLTLIIFDD160 pKa = 3.97DD161 pKa = 4.06EE162 pKa = 5.1KK163 pKa = 10.96YY164 pKa = 10.59QSLIPNYY171 pKa = 8.93INQLIAYY178 pKa = 5.86TQPHH182 pKa = 7.71IIPTWQGIADD192 pKa = 4.32FSDD195 pKa = 3.56TYY197 pKa = 10.93LRR199 pKa = 11.84SYY201 pKa = 10.09FKK203 pKa = 10.89RR204 pKa = 11.84PFEE207 pKa = 4.06LTASNLAAPQKK218 pKa = 10.75HH219 pKa = 5.88NLSPMTRR226 pKa = 11.84SIFNNTGRR234 pKa = 11.84EE235 pKa = 3.82DD236 pKa = 3.4AVIRR240 pKa = 11.84KK241 pKa = 9.45LYY243 pKa = 10.72GYY245 pKa = 10.27GEE247 pKa = 4.44YY248 pKa = 11.12VFIKK252 pKa = 11.0YY253 pKa = 8.71EE254 pKa = 4.09GCLITWTGIYY264 pKa = 10.78GEE266 pKa = 4.2VTMMVNLSKK275 pKa = 10.66RR276 pKa = 11.84DD277 pKa = 3.55LGLDD281 pKa = 3.25VGDD284 pKa = 5.72DD285 pKa = 3.58YY286 pKa = 11.97LKK288 pKa = 10.45EE289 pKa = 4.1YY290 pKa = 10.77KK291 pKa = 10.13KK292 pKa = 10.91LLFYY296 pKa = 10.92GVITDD301 pKa = 5.76AIPSGISTRR310 pKa = 11.84STIMKK315 pKa = 9.91ISPHH319 pKa = 7.59KK320 pKa = 10.14MMNPSGGALAVLSKK334 pKa = 10.58FLEE337 pKa = 4.48AVVSTNVINATLVVYY352 pKa = 10.51AEE354 pKa = 4.34KK355 pKa = 10.85GAGKK359 pKa = 8.68TSFLSTYY366 pKa = 10.93AEE368 pKa = 4.08QLSLASGQVVGHH380 pKa = 6.99LSSDD384 pKa = 3.14AYY386 pKa = 10.61GRR388 pKa = 11.84WLAKK392 pKa = 9.89TKK394 pKa = 10.46DD395 pKa = 3.21IEE397 pKa = 4.52EE398 pKa = 4.17PSFAYY403 pKa = 10.15DD404 pKa = 3.46YY405 pKa = 11.4VLSLDD410 pKa = 3.57TDD412 pKa = 4.2DD413 pKa = 4.63NEE415 pKa = 4.92SYY417 pKa = 10.91YY418 pKa = 9.29EE419 pKa = 4.04QKK421 pKa = 10.93ASEE424 pKa = 4.65LLMSHH429 pKa = 7.16GISEE433 pKa = 4.36VAQYY437 pKa = 11.15EE438 pKa = 4.28LLSVRR443 pKa = 11.84KK444 pKa = 8.85KK445 pKa = 10.38IKK447 pKa = 10.4MMDD450 pKa = 3.36EE451 pKa = 4.0MNEE454 pKa = 3.76VLIAQLEE461 pKa = 4.3NADD464 pKa = 3.67THH466 pKa = 6.13SEE468 pKa = 4.04RR469 pKa = 11.84NFYY472 pKa = 11.58YY473 pKa = 9.9MVSTGKK479 pKa = 6.19TTPRR483 pKa = 11.84ILIVEE488 pKa = 4.06GHH490 pKa = 6.41FNAQDD495 pKa = 3.26ATIARR500 pKa = 11.84TDD502 pKa = 3.41TTVLLRR508 pKa = 11.84TINDD512 pKa = 3.35TTQAMRR518 pKa = 11.84DD519 pKa = 3.56RR520 pKa = 11.84QRR522 pKa = 11.84GGVVQLFLRR531 pKa = 11.84DD532 pKa = 3.14TYY534 pKa = 11.61YY535 pKa = 11.31RR536 pKa = 11.84LLPALHH542 pKa = 5.25TTVYY546 pKa = 8.96PFEE549 pKa = 4.09MLEE552 pKa = 4.54SIKK555 pKa = 10.15RR556 pKa = 11.84WKK558 pKa = 8.44WVHH561 pKa = 5.43
Molecular weight: 63.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8294 |
248 |
1333 |
754.0 |
85.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.86 ± 0.476 | 0.699 ± 0.177 |
5.836 ± 0.22 | 5.45 ± 0.318 |
3.786 ± 0.208 | 4.702 ± 0.253 |
2.64 ± 0.119 | 6.788 ± 0.264 |
4.292 ± 0.404 | 8.934 ± 0.387 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.894 ± 0.19 | 5.619 ± 0.242 |
4.365 ± 0.364 | 3.81 ± 0.2 |
6.004 ± 0.273 | 7.801 ± 0.322 |
7.403 ± 0.395 | 6.933 ± 0.353 |
0.796 ± 0.088 | 4.389 ± 0.363 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
