
Bifidobacterium felsineum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Bifidobacteriales; Bifidobacteriaceae; Bifidobacterium
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
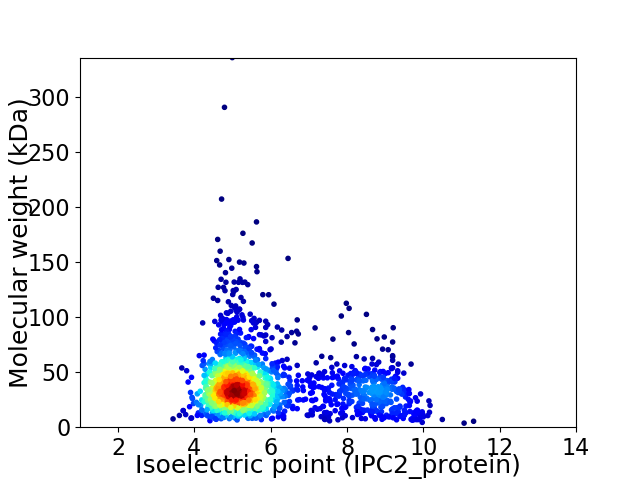
Virtual 2D-PAGE plot for 1828 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2M9HLG2|A0A2M9HLG2_9BIFI Sucrose phosphorylase OS=Bifidobacterium felsineum OX=2045440 GN=CSQ86_00730 PE=3 SV=1
MM1 pKa = 7.21TLEE4 pKa = 4.45NSDD7 pKa = 3.49AAQALVQEE15 pKa = 4.54IVRR18 pKa = 11.84RR19 pKa = 11.84IVRR22 pKa = 11.84DD23 pKa = 3.49TSKK26 pKa = 9.95MAVSFTPEE34 pKa = 3.72LDD36 pKa = 3.37VRR38 pKa = 11.84PDD40 pKa = 3.35LLGNFIGGDD49 pKa = 4.28SYY51 pKa = 11.23WSPQGVPVTSADD63 pKa = 3.51GSPLYY68 pKa = 10.21PLMQIRR74 pKa = 11.84CTEE77 pKa = 4.07PSLPGYY83 pKa = 6.29PTRR86 pKa = 11.84GLLQFFIGTDD96 pKa = 2.84IMYY99 pKa = 10.36GCDD102 pKa = 3.49LSADD106 pKa = 3.91YY107 pKa = 10.96DD108 pKa = 4.17SEE110 pKa = 5.21ADD112 pKa = 3.31NDD114 pKa = 3.97NRR116 pKa = 11.84CVRR119 pKa = 11.84YY120 pKa = 9.56IPEE123 pKa = 4.18VPSVLSGAEE132 pKa = 4.07IVRR135 pKa = 11.84LPHH138 pKa = 6.86IGADD142 pKa = 3.67SDD144 pKa = 4.48PDD146 pKa = 3.22LHH148 pKa = 6.6TPFRR152 pKa = 11.84SNFHH156 pKa = 5.86GCALRR161 pKa = 11.84PALIEE166 pKa = 4.54DD167 pKa = 4.39PVSHH171 pKa = 6.64EE172 pKa = 4.1CDD174 pKa = 3.31EE175 pKa = 4.6FEE177 pKa = 4.37PAYY180 pKa = 10.12EE181 pKa = 4.15SIIAEE186 pKa = 4.31FDD188 pKa = 3.56PAVIEE193 pKa = 4.13ATEE196 pKa = 4.17DD197 pKa = 3.37AVGDD201 pKa = 4.03LDD203 pKa = 4.61QAVMDD208 pKa = 4.12QLNAVSPGSWFQMGGHH224 pKa = 6.59PVFTQSDD231 pKa = 3.57VRR233 pKa = 11.84EE234 pKa = 4.65DD235 pKa = 3.88YY236 pKa = 11.25DD237 pKa = 5.59DD238 pKa = 4.39EE239 pKa = 5.25LVLLKK244 pKa = 10.4VDD246 pKa = 4.07SYY248 pKa = 12.04DD249 pKa = 4.69DD250 pKa = 3.47VLMWGDD256 pKa = 3.61SGCANFLIRR265 pKa = 11.84RR266 pKa = 11.84EE267 pKa = 4.18DD268 pKa = 3.66LEE270 pKa = 4.04QLDD273 pKa = 4.64FSHH276 pKa = 7.29VLYY279 pKa = 9.67TWDD282 pKa = 3.85CLL284 pKa = 3.89
MM1 pKa = 7.21TLEE4 pKa = 4.45NSDD7 pKa = 3.49AAQALVQEE15 pKa = 4.54IVRR18 pKa = 11.84RR19 pKa = 11.84IVRR22 pKa = 11.84DD23 pKa = 3.49TSKK26 pKa = 9.95MAVSFTPEE34 pKa = 3.72LDD36 pKa = 3.37VRR38 pKa = 11.84PDD40 pKa = 3.35LLGNFIGGDD49 pKa = 4.28SYY51 pKa = 11.23WSPQGVPVTSADD63 pKa = 3.51GSPLYY68 pKa = 10.21PLMQIRR74 pKa = 11.84CTEE77 pKa = 4.07PSLPGYY83 pKa = 6.29PTRR86 pKa = 11.84GLLQFFIGTDD96 pKa = 2.84IMYY99 pKa = 10.36GCDD102 pKa = 3.49LSADD106 pKa = 3.91YY107 pKa = 10.96DD108 pKa = 4.17SEE110 pKa = 5.21ADD112 pKa = 3.31NDD114 pKa = 3.97NRR116 pKa = 11.84CVRR119 pKa = 11.84YY120 pKa = 9.56IPEE123 pKa = 4.18VPSVLSGAEE132 pKa = 4.07IVRR135 pKa = 11.84LPHH138 pKa = 6.86IGADD142 pKa = 3.67SDD144 pKa = 4.48PDD146 pKa = 3.22LHH148 pKa = 6.6TPFRR152 pKa = 11.84SNFHH156 pKa = 5.86GCALRR161 pKa = 11.84PALIEE166 pKa = 4.54DD167 pKa = 4.39PVSHH171 pKa = 6.64EE172 pKa = 4.1CDD174 pKa = 3.31EE175 pKa = 4.6FEE177 pKa = 4.37PAYY180 pKa = 10.12EE181 pKa = 4.15SIIAEE186 pKa = 4.31FDD188 pKa = 3.56PAVIEE193 pKa = 4.13ATEE196 pKa = 4.17DD197 pKa = 3.37AVGDD201 pKa = 4.03LDD203 pKa = 4.61QAVMDD208 pKa = 4.12QLNAVSPGSWFQMGGHH224 pKa = 6.59PVFTQSDD231 pKa = 3.57VRR233 pKa = 11.84EE234 pKa = 4.65DD235 pKa = 3.88YY236 pKa = 11.25DD237 pKa = 5.59DD238 pKa = 4.39EE239 pKa = 5.25LVLLKK244 pKa = 10.4VDD246 pKa = 4.07SYY248 pKa = 12.04DD249 pKa = 4.69DD250 pKa = 3.47VLMWGDD256 pKa = 3.61SGCANFLIRR265 pKa = 11.84RR266 pKa = 11.84EE267 pKa = 4.18DD268 pKa = 3.66LEE270 pKa = 4.04QLDD273 pKa = 4.64FSHH276 pKa = 7.29VLYY279 pKa = 9.67TWDD282 pKa = 3.85CLL284 pKa = 3.89
Molecular weight: 31.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2M9HLA6|A0A2M9HLA6_9BIFI Metallophos domain-containing protein OS=Bifidobacterium felsineum OX=2045440 GN=CSQ86_00360 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 4.82MKK15 pKa = 9.64HH16 pKa = 5.7GFRR19 pKa = 11.84LRR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84SGRR28 pKa = 11.84AVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.06GRR39 pKa = 11.84KK40 pKa = 8.87SLSAA44 pKa = 3.86
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 4.82MKK15 pKa = 9.64HH16 pKa = 5.7GFRR19 pKa = 11.84LRR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84SGRR28 pKa = 11.84AVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.06GRR39 pKa = 11.84KK40 pKa = 8.87SLSAA44 pKa = 3.86
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
685830 |
29 |
3163 |
375.2 |
40.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.082 ± 0.078 | 0.861 ± 0.018 |
6.276 ± 0.048 | 5.668 ± 0.052 |
3.594 ± 0.033 | 7.878 ± 0.046 |
2.192 ± 0.03 | 5.665 ± 0.041 |
4.228 ± 0.047 | 8.855 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.548 ± 0.028 | 3.638 ± 0.044 |
4.556 ± 0.036 | 3.522 ± 0.036 |
5.246 ± 0.057 | 6.092 ± 0.051 |
6.188 ± 0.052 | 7.699 ± 0.05 |
1.392 ± 0.025 | 2.823 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
