
Sulfolobus turreted icosahedral virus 2
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Belfryvirales; Turriviridae; Alphaturrivirus
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
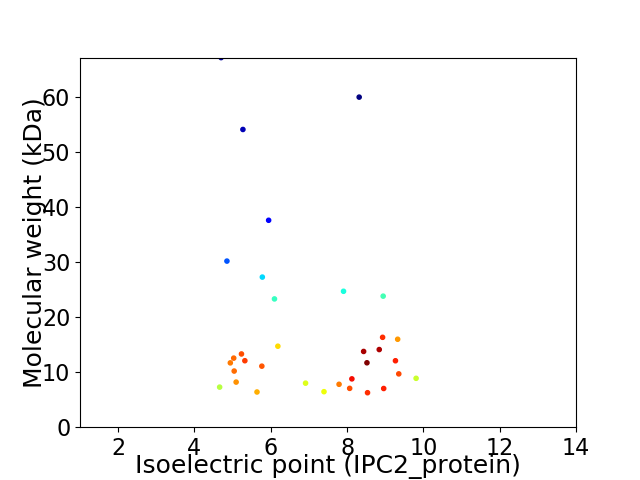
Virtual 2D-PAGE plot for 34 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D5IEY3|D5IEY3_9VIRU Uncharacterized protein OS=Sulfolobus turreted icosahedral virus 2 OX=754004 GN=STIV2_B67 PE=4 SV=1
MM1 pKa = 8.24DD2 pKa = 4.09EE3 pKa = 4.11FKK5 pKa = 10.7EE6 pKa = 4.21VKK8 pKa = 10.03EE9 pKa = 3.75KK10 pKa = 10.77FEE12 pKa = 4.16RR13 pKa = 11.84YY14 pKa = 9.61KK15 pKa = 10.95FDD17 pKa = 2.85VEE19 pKa = 4.48YY20 pKa = 10.29IDD22 pKa = 5.41RR23 pKa = 11.84EE24 pKa = 4.15YY25 pKa = 10.61TINANYY31 pKa = 8.84PKK33 pKa = 10.68AEE35 pKa = 3.93IPIGEE40 pKa = 4.16RR41 pKa = 11.84NGFTGIVVNNGTAILTITLVKK62 pKa = 9.99VAHH65 pKa = 6.84ASTTADD71 pKa = 3.61LGVSTSDD78 pKa = 2.85VGTIILAPGQVLQLTNFPLNEE99 pKa = 3.64IKK101 pKa = 10.53AVYY104 pKa = 9.6NPNSPSSASSVVQVKK119 pKa = 10.01GVIHH123 pKa = 6.75PLFEE127 pKa = 4.47VPVKK131 pKa = 11.04LNVTDD136 pKa = 3.68IVSIQQSNEE145 pKa = 3.48NLLITTNTTINGGNYY160 pKa = 9.58NDD162 pKa = 5.12IVIQPNVTVNFDD174 pKa = 3.71GFVTAKK180 pKa = 10.12KK181 pKa = 10.27IIMQSGSTVNVNGVLIVGEE200 pKa = 4.34IEE202 pKa = 4.11ADD204 pKa = 3.68SPATLASSTPPINLTVNGVLVANYY228 pKa = 8.72VQSDD232 pKa = 4.18VNVEE236 pKa = 4.09VASGFMGVKK245 pKa = 8.39TLXYY249 pKa = 9.48NGQVLYY255 pKa = 11.01VNQGAFLFADD265 pKa = 3.5TMIFSNNLNSLTYY278 pKa = 10.25NSSISSAPGNIFANEE293 pKa = 4.01IYY295 pKa = 10.68FEE297 pKa = 4.34TLLSQQLSVNMNITAKK313 pKa = 9.86KK314 pKa = 9.57IRR316 pKa = 11.84MGGLMGGLMEE326 pKa = 5.41IMAGLLTSLSISNLQFAINGNNLIIKK352 pKa = 7.33MKK354 pKa = 9.93KK355 pKa = 10.22AIIFGFNANVNNSTIEE371 pKa = 3.95IEE373 pKa = 4.21DD374 pKa = 4.1SIIASPSFYY383 pKa = 10.33AYY385 pKa = 10.19SGSSTIYY392 pKa = 10.34IIIKK396 pKa = 10.38GNTQIYY402 pKa = 8.94QLSINSSLTSYY413 pKa = 10.78IFFRR417 pKa = 11.84QNEE420 pKa = 4.1NGQGNIYY427 pKa = 10.16LYY429 pKa = 9.98NASFATSTYY438 pKa = 8.1WRR440 pKa = 11.84IDD442 pKa = 3.3FGVTVTAYY450 pKa = 7.91PASFSTSSSNSITVNGQLYY469 pKa = 9.2MSSNLTTTSASSTTSSSVTIGLPFTVTGFGIAIISQNTNASANQGGYY516 pKa = 10.1LAGNTIQMNNLQLSKK531 pKa = 10.91GSYY534 pKa = 8.67STTTQTGSGNNTYY547 pKa = 9.94WYY549 pKa = 7.69ITSIQGSAPGLYY561 pKa = 10.17YY562 pKa = 10.77LGVYY566 pKa = 10.43DD567 pKa = 4.24EE568 pKa = 5.04TSGATNYY575 pKa = 9.6IAEE578 pKa = 4.18IVVYY582 pKa = 10.47VGTVNQYY589 pKa = 11.38ALSNPLGFVAEE600 pKa = 4.55GGVADD605 pKa = 4.17TSGNTYY611 pKa = 10.83AILNSSASAGTITLSGLAAVV631 pKa = 4.65
MM1 pKa = 8.24DD2 pKa = 4.09EE3 pKa = 4.11FKK5 pKa = 10.7EE6 pKa = 4.21VKK8 pKa = 10.03EE9 pKa = 3.75KK10 pKa = 10.77FEE12 pKa = 4.16RR13 pKa = 11.84YY14 pKa = 9.61KK15 pKa = 10.95FDD17 pKa = 2.85VEE19 pKa = 4.48YY20 pKa = 10.29IDD22 pKa = 5.41RR23 pKa = 11.84EE24 pKa = 4.15YY25 pKa = 10.61TINANYY31 pKa = 8.84PKK33 pKa = 10.68AEE35 pKa = 3.93IPIGEE40 pKa = 4.16RR41 pKa = 11.84NGFTGIVVNNGTAILTITLVKK62 pKa = 9.99VAHH65 pKa = 6.84ASTTADD71 pKa = 3.61LGVSTSDD78 pKa = 2.85VGTIILAPGQVLQLTNFPLNEE99 pKa = 3.64IKK101 pKa = 10.53AVYY104 pKa = 9.6NPNSPSSASSVVQVKK119 pKa = 10.01GVIHH123 pKa = 6.75PLFEE127 pKa = 4.47VPVKK131 pKa = 11.04LNVTDD136 pKa = 3.68IVSIQQSNEE145 pKa = 3.48NLLITTNTTINGGNYY160 pKa = 9.58NDD162 pKa = 5.12IVIQPNVTVNFDD174 pKa = 3.71GFVTAKK180 pKa = 10.12KK181 pKa = 10.27IIMQSGSTVNVNGVLIVGEE200 pKa = 4.34IEE202 pKa = 4.11ADD204 pKa = 3.68SPATLASSTPPINLTVNGVLVANYY228 pKa = 8.72VQSDD232 pKa = 4.18VNVEE236 pKa = 4.09VASGFMGVKK245 pKa = 8.39TLXYY249 pKa = 9.48NGQVLYY255 pKa = 11.01VNQGAFLFADD265 pKa = 3.5TMIFSNNLNSLTYY278 pKa = 10.25NSSISSAPGNIFANEE293 pKa = 4.01IYY295 pKa = 10.68FEE297 pKa = 4.34TLLSQQLSVNMNITAKK313 pKa = 9.86KK314 pKa = 9.57IRR316 pKa = 11.84MGGLMGGLMEE326 pKa = 5.41IMAGLLTSLSISNLQFAINGNNLIIKK352 pKa = 7.33MKK354 pKa = 9.93KK355 pKa = 10.22AIIFGFNANVNNSTIEE371 pKa = 3.95IEE373 pKa = 4.21DD374 pKa = 4.1SIIASPSFYY383 pKa = 10.33AYY385 pKa = 10.19SGSSTIYY392 pKa = 10.34IIIKK396 pKa = 10.38GNTQIYY402 pKa = 8.94QLSINSSLTSYY413 pKa = 10.78IFFRR417 pKa = 11.84QNEE420 pKa = 4.1NGQGNIYY427 pKa = 10.16LYY429 pKa = 9.98NASFATSTYY438 pKa = 8.1WRR440 pKa = 11.84IDD442 pKa = 3.3FGVTVTAYY450 pKa = 7.91PASFSTSSSNSITVNGQLYY469 pKa = 9.2MSSNLTTTSASSTTSSSVTIGLPFTVTGFGIAIISQNTNASANQGGYY516 pKa = 10.1LAGNTIQMNNLQLSKK531 pKa = 10.91GSYY534 pKa = 8.67STTTQTGSGNNTYY547 pKa = 9.94WYY549 pKa = 7.69ITSIQGSAPGLYY561 pKa = 10.17YY562 pKa = 10.77LGVYY566 pKa = 10.43DD567 pKa = 4.24EE568 pKa = 5.04TSGATNYY575 pKa = 9.6IAEE578 pKa = 4.18IVVYY582 pKa = 10.47VGTVNQYY589 pKa = 11.38ALSNPLGFVAEE600 pKa = 4.55GGVADD605 pKa = 4.17TSGNTYY611 pKa = 10.83AILNSSASAGTITLSGLAAVV631 pKa = 4.65
Molecular weight: 67.12 kDa
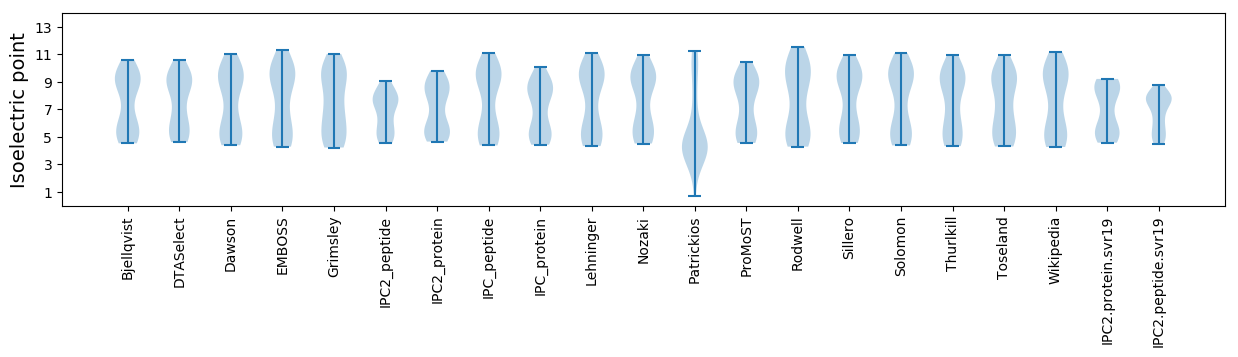
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D5IEX4|D5IEX4_9VIRU Uncharacterized protein OS=Sulfolobus turreted icosahedral virus 2 OX=754004 GN=STIV2_C54a PE=4 SV=1
MM1 pKa = 7.26AKK3 pKa = 9.94KK4 pKa = 9.68WIQKK8 pKa = 9.65AIKK11 pKa = 9.9HH12 pKa = 5.58KK13 pKa = 10.63GALKK17 pKa = 9.41EE18 pKa = 4.22WIKK21 pKa = 10.93KK22 pKa = 7.08EE23 pKa = 3.99HH24 pKa = 6.14PTLLRR29 pKa = 11.84KK30 pKa = 10.35DD31 pKa = 3.74GEE33 pKa = 3.98ISITKK38 pKa = 10.31LEE40 pKa = 4.02EE41 pKa = 4.08FYY43 pKa = 10.87RR44 pKa = 11.84KK45 pKa = 9.78HH46 pKa = 7.0KK47 pKa = 10.95DD48 pKa = 2.89EE49 pKa = 4.5LTPHH53 pKa = 7.49RR54 pKa = 11.84KK55 pKa = 8.92RR56 pKa = 11.84QILLALRR63 pKa = 11.84LYY65 pKa = 10.86RR66 pKa = 11.84MRR68 pKa = 11.84RR69 pKa = 11.84KK70 pKa = 10.13KK71 pKa = 10.55SS72 pKa = 3.12
MM1 pKa = 7.26AKK3 pKa = 9.94KK4 pKa = 9.68WIQKK8 pKa = 9.65AIKK11 pKa = 9.9HH12 pKa = 5.58KK13 pKa = 10.63GALKK17 pKa = 9.41EE18 pKa = 4.22WIKK21 pKa = 10.93KK22 pKa = 7.08EE23 pKa = 3.99HH24 pKa = 6.14PTLLRR29 pKa = 11.84KK30 pKa = 10.35DD31 pKa = 3.74GEE33 pKa = 3.98ISITKK38 pKa = 10.31LEE40 pKa = 4.02EE41 pKa = 4.08FYY43 pKa = 10.87RR44 pKa = 11.84KK45 pKa = 9.78HH46 pKa = 7.0KK47 pKa = 10.95DD48 pKa = 2.89EE49 pKa = 4.5LTPHH53 pKa = 7.49RR54 pKa = 11.84KK55 pKa = 8.92RR56 pKa = 11.84QILLALRR63 pKa = 11.84LYY65 pKa = 10.86RR66 pKa = 11.84MRR68 pKa = 11.84RR69 pKa = 11.84KK70 pKa = 10.13KK71 pKa = 10.55SS72 pKa = 3.12
Molecular weight: 8.88 kDa
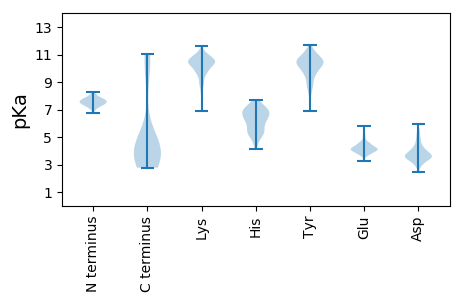
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5395 |
54 |
631 |
158.7 |
17.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.228 ± 0.556 | 0.686 ± 0.177 |
4.133 ± 0.326 | 7.804 ± 0.905 |
4.133 ± 0.438 | 5.876 ± 0.559 |
1.538 ± 0.214 | 9.027 ± 0.544 |
8.378 ± 1.194 | 10.361 ± 0.467 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.909 ± 0.176 | 5.042 ± 0.637 |
3.911 ± 0.543 | 3.207 ± 0.298 |
4.059 ± 0.441 | 7.451 ± 0.779 |
5.338 ± 0.608 | 5.82 ± 0.445 |
0.704 ± 0.168 | 4.374 ± 0.355 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
