
Tapara virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Tapara phlebovirus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
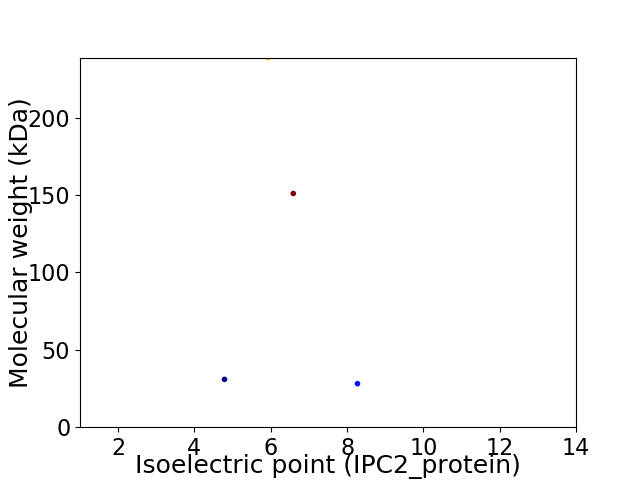
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S5SHW6|A0A1S5SHW6_9VIRU Nonstructural protein OS=Tapara virus OX=1926501 PE=4 SV=1
MM1 pKa = 7.82SSYY4 pKa = 10.97LYY6 pKa = 10.12DD7 pKa = 3.65RR8 pKa = 11.84PIIFRR13 pKa = 11.84QSQEE17 pKa = 4.12DD18 pKa = 4.13CSVQYY23 pKa = 11.02LAFNSFTDD31 pKa = 4.7RR32 pKa = 11.84EE33 pKa = 4.51VCVNNDD39 pKa = 2.96MEE41 pKa = 4.93IPVEE45 pKa = 4.32NYY47 pKa = 7.4MASCAFRR54 pKa = 11.84EE55 pKa = 4.26SLQDD59 pKa = 3.71FYY61 pKa = 11.6RR62 pKa = 11.84AGTLPLKK69 pKa = 10.08WGDD72 pKa = 3.57EE73 pKa = 4.24MPVNRR78 pKa = 11.84VTKK81 pKa = 10.32GSYY84 pKa = 10.25RR85 pKa = 11.84LMDD88 pKa = 3.66GVIEE92 pKa = 4.17SLSRR96 pKa = 11.84FSRR99 pKa = 11.84EE100 pKa = 4.31DD101 pKa = 3.05IIKK104 pKa = 10.35SYY106 pKa = 10.73LPNVQNALSWPLGYY120 pKa = 7.68PTLDD124 pKa = 3.91FIKK127 pKa = 10.19ACVMDD132 pKa = 3.87TPLYY136 pKa = 8.61TRR138 pKa = 11.84KK139 pKa = 8.48STHH142 pKa = 5.38ATLIFRR148 pKa = 11.84TGQPADD154 pKa = 4.17CLDD157 pKa = 3.4QSFVNSHH164 pKa = 5.49RR165 pKa = 11.84RR166 pKa = 11.84IVMEE170 pKa = 3.88SVTRR174 pKa = 11.84GFDD177 pKa = 3.09VKK179 pKa = 10.3TFTGQNLVRR188 pKa = 11.84DD189 pKa = 4.14IASLQCVRR197 pKa = 11.84VLNAYY202 pKa = 9.45QADD205 pKa = 4.03LLYY208 pKa = 10.93CGVSTEE214 pKa = 4.18LTKK217 pKa = 10.67EE218 pKa = 3.9LSKK221 pKa = 11.06LRR223 pKa = 11.84IIHH226 pKa = 7.12DD227 pKa = 3.64GHH229 pKa = 6.42PQNPLGNQRR238 pKa = 11.84WIASPDD244 pKa = 3.02SWDD247 pKa = 3.45YY248 pKa = 11.41RR249 pKa = 11.84PNPDD253 pKa = 3.78VEE255 pKa = 5.23LDD257 pKa = 3.46FDD259 pKa = 5.65LDD261 pKa = 3.62ISDD264 pKa = 4.13SSEE267 pKa = 4.12DD268 pKa = 3.59EE269 pKa = 4.07
MM1 pKa = 7.82SSYY4 pKa = 10.97LYY6 pKa = 10.12DD7 pKa = 3.65RR8 pKa = 11.84PIIFRR13 pKa = 11.84QSQEE17 pKa = 4.12DD18 pKa = 4.13CSVQYY23 pKa = 11.02LAFNSFTDD31 pKa = 4.7RR32 pKa = 11.84EE33 pKa = 4.51VCVNNDD39 pKa = 2.96MEE41 pKa = 4.93IPVEE45 pKa = 4.32NYY47 pKa = 7.4MASCAFRR54 pKa = 11.84EE55 pKa = 4.26SLQDD59 pKa = 3.71FYY61 pKa = 11.6RR62 pKa = 11.84AGTLPLKK69 pKa = 10.08WGDD72 pKa = 3.57EE73 pKa = 4.24MPVNRR78 pKa = 11.84VTKK81 pKa = 10.32GSYY84 pKa = 10.25RR85 pKa = 11.84LMDD88 pKa = 3.66GVIEE92 pKa = 4.17SLSRR96 pKa = 11.84FSRR99 pKa = 11.84EE100 pKa = 4.31DD101 pKa = 3.05IIKK104 pKa = 10.35SYY106 pKa = 10.73LPNVQNALSWPLGYY120 pKa = 7.68PTLDD124 pKa = 3.91FIKK127 pKa = 10.19ACVMDD132 pKa = 3.87TPLYY136 pKa = 8.61TRR138 pKa = 11.84KK139 pKa = 8.48STHH142 pKa = 5.38ATLIFRR148 pKa = 11.84TGQPADD154 pKa = 4.17CLDD157 pKa = 3.4QSFVNSHH164 pKa = 5.49RR165 pKa = 11.84RR166 pKa = 11.84IVMEE170 pKa = 3.88SVTRR174 pKa = 11.84GFDD177 pKa = 3.09VKK179 pKa = 10.3TFTGQNLVRR188 pKa = 11.84DD189 pKa = 4.14IASLQCVRR197 pKa = 11.84VLNAYY202 pKa = 9.45QADD205 pKa = 4.03LLYY208 pKa = 10.93CGVSTEE214 pKa = 4.18LTKK217 pKa = 10.67EE218 pKa = 3.9LSKK221 pKa = 11.06LRR223 pKa = 11.84IIHH226 pKa = 7.12DD227 pKa = 3.64GHH229 pKa = 6.42PQNPLGNQRR238 pKa = 11.84WIASPDD244 pKa = 3.02SWDD247 pKa = 3.45YY248 pKa = 11.41RR249 pKa = 11.84PNPDD253 pKa = 3.78VEE255 pKa = 5.23LDD257 pKa = 3.46FDD259 pKa = 5.65LDD261 pKa = 3.62ISDD264 pKa = 4.13SSEE267 pKa = 4.12DD268 pKa = 3.59EE269 pKa = 4.07
Molecular weight: 30.89 kDa
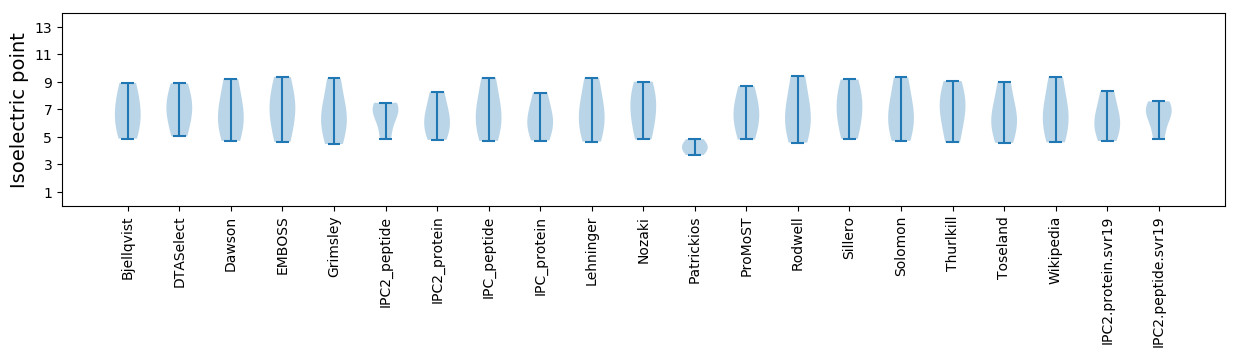
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S5SHW6|A0A1S5SHW6_9VIRU Nonstructural protein OS=Tapara virus OX=1926501 PE=4 SV=1
MM1 pKa = 7.85SDD3 pKa = 3.66YY4 pKa = 10.91EE5 pKa = 4.1KK6 pKa = 11.01LAIEE10 pKa = 4.28LQGEE14 pKa = 4.54PIDD17 pKa = 4.04KK18 pKa = 10.55NAIQRR23 pKa = 11.84WMSEE27 pKa = 3.61FAYY30 pKa = 10.28EE31 pKa = 4.76GFDD34 pKa = 3.17AANIVKK40 pKa = 10.08LVHH43 pKa = 6.01EE44 pKa = 4.37RR45 pKa = 11.84AKK47 pKa = 10.89AAGRR51 pKa = 11.84DD52 pKa = 3.13WKK54 pKa = 11.16LDD56 pKa = 3.6VKK58 pKa = 11.23KK59 pKa = 10.36MIVLALTRR67 pKa = 11.84GNKK70 pKa = 7.3VDD72 pKa = 4.16KK73 pKa = 9.79MIKK76 pKa = 10.1KK77 pKa = 9.36MSEE80 pKa = 3.39KK81 pKa = 10.4GAATVTEE88 pKa = 4.22LVRR91 pKa = 11.84VYY93 pKa = 10.58SLKK96 pKa = 10.82SGNPTRR102 pKa = 11.84DD103 pKa = 3.59DD104 pKa = 3.55LTLSRR109 pKa = 11.84VAAAFAGYY117 pKa = 7.77TCQALPHH124 pKa = 6.35LAEE127 pKa = 5.64AIPVSGSQMDD137 pKa = 4.08EE138 pKa = 3.94VVKK141 pKa = 10.78NYY143 pKa = 7.42PRR145 pKa = 11.84PMMHH149 pKa = 7.17PAFSGLIDD157 pKa = 3.48PTFKK161 pKa = 11.22EE162 pKa = 4.13EE163 pKa = 4.39TKK165 pKa = 10.67DD166 pKa = 3.5ALINAHH172 pKa = 5.93SLFMYY177 pKa = 10.33HH178 pKa = 5.83FTVQINEE185 pKa = 3.93KK186 pKa = 9.66MRR188 pKa = 11.84GKK190 pKa = 9.96SDD192 pKa = 3.38KK193 pKa = 10.7EE194 pKa = 4.14IISSFQQPMYY204 pKa = 10.88AAINSGFMKK213 pKa = 10.74GEE215 pKa = 3.8TRR217 pKa = 11.84RR218 pKa = 11.84AMLRR222 pKa = 11.84TLGIVDD228 pKa = 4.38ANWVPSSIVQEE239 pKa = 3.63AAKK242 pKa = 10.01IYY244 pKa = 9.91KK245 pKa = 9.78EE246 pKa = 4.31KK247 pKa = 11.41YY248 pKa = 9.27NDD250 pKa = 3.2
MM1 pKa = 7.85SDD3 pKa = 3.66YY4 pKa = 10.91EE5 pKa = 4.1KK6 pKa = 11.01LAIEE10 pKa = 4.28LQGEE14 pKa = 4.54PIDD17 pKa = 4.04KK18 pKa = 10.55NAIQRR23 pKa = 11.84WMSEE27 pKa = 3.61FAYY30 pKa = 10.28EE31 pKa = 4.76GFDD34 pKa = 3.17AANIVKK40 pKa = 10.08LVHH43 pKa = 6.01EE44 pKa = 4.37RR45 pKa = 11.84AKK47 pKa = 10.89AAGRR51 pKa = 11.84DD52 pKa = 3.13WKK54 pKa = 11.16LDD56 pKa = 3.6VKK58 pKa = 11.23KK59 pKa = 10.36MIVLALTRR67 pKa = 11.84GNKK70 pKa = 7.3VDD72 pKa = 4.16KK73 pKa = 9.79MIKK76 pKa = 10.1KK77 pKa = 9.36MSEE80 pKa = 3.39KK81 pKa = 10.4GAATVTEE88 pKa = 4.22LVRR91 pKa = 11.84VYY93 pKa = 10.58SLKK96 pKa = 10.82SGNPTRR102 pKa = 11.84DD103 pKa = 3.59DD104 pKa = 3.55LTLSRR109 pKa = 11.84VAAAFAGYY117 pKa = 7.77TCQALPHH124 pKa = 6.35LAEE127 pKa = 5.64AIPVSGSQMDD137 pKa = 4.08EE138 pKa = 3.94VVKK141 pKa = 10.78NYY143 pKa = 7.42PRR145 pKa = 11.84PMMHH149 pKa = 7.17PAFSGLIDD157 pKa = 3.48PTFKK161 pKa = 11.22EE162 pKa = 4.13EE163 pKa = 4.39TKK165 pKa = 10.67DD166 pKa = 3.5ALINAHH172 pKa = 5.93SLFMYY177 pKa = 10.33HH178 pKa = 5.83FTVQINEE185 pKa = 3.93KK186 pKa = 9.66MRR188 pKa = 11.84GKK190 pKa = 9.96SDD192 pKa = 3.38KK193 pKa = 10.7EE194 pKa = 4.14IISSFQQPMYY204 pKa = 10.88AAINSGFMKK213 pKa = 10.74GEE215 pKa = 3.8TRR217 pKa = 11.84RR218 pKa = 11.84AMLRR222 pKa = 11.84TLGIVDD228 pKa = 4.38ANWVPSSIVQEE239 pKa = 3.63AAKK242 pKa = 10.01IYY244 pKa = 9.91KK245 pKa = 9.78EE246 pKa = 4.31KK247 pKa = 11.41YY248 pKa = 9.27NDD250 pKa = 3.2
Molecular weight: 28.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3951 |
250 |
2089 |
987.8 |
112.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.961 ± 0.835 | 2.455 ± 0.826 |
5.695 ± 0.637 | 7.669 ± 0.57 |
4.885 ± 0.365 | 5.669 ± 0.335 |
2.556 ± 0.247 | 6.96 ± 0.388 |
7.213 ± 0.731 | 8.226 ± 0.273 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.265 ± 0.361 | 3.872 ± 0.496 |
3.569 ± 0.394 | 2.784 ± 0.374 |
5.442 ± 0.313 | 9.415 ± 0.446 |
5.29 ± 0.474 | 5.897 ± 0.194 |
1.215 ± 0.06 | 2.961 ± 0.351 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
