
Rhynchosia yellow mosaic virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 8.32
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
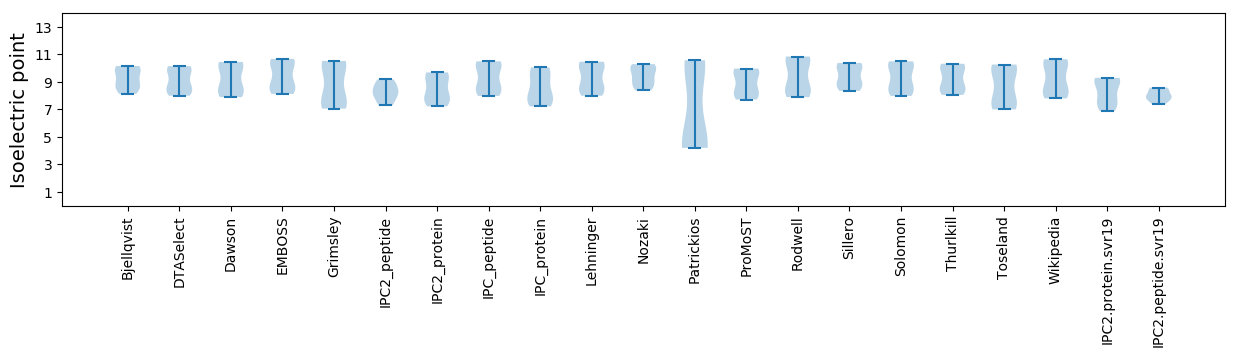
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|C8TES4|C8TES4_9GEMI Movement protein BC1 OS=Rhynchosia yellow mosaic virus OX=529680 GN=bc1 PE=3 SV=2
MM1 pKa = 8.04DD2 pKa = 5.02NYY4 pKa = 11.14SGAVVNNKK12 pKa = 9.82YY13 pKa = 10.02IEE15 pKa = 4.33SKK17 pKa = 9.13RR18 pKa = 11.84CEE20 pKa = 3.84YY21 pKa = 10.88RR22 pKa = 11.84LTNNEE27 pKa = 4.22TPIMLQFPSSLEE39 pKa = 3.87QTKK42 pKa = 10.0VRR44 pKa = 11.84MLGKK48 pKa = 10.07CMKK51 pKa = 9.46VDD53 pKa = 4.32HH54 pKa = 6.68IVIEE58 pKa = 4.22YY59 pKa = 10.2RR60 pKa = 11.84NQVPFNATGSVIVTIRR76 pKa = 11.84DD77 pKa = 3.21TRR79 pKa = 11.84LSHH82 pKa = 5.99EE83 pKa = 4.3QAAQAAFTFPIACNVDD99 pKa = 2.84LHH101 pKa = 6.36YY102 pKa = 10.7FSSSFFSLKK111 pKa = 10.84DD112 pKa = 3.25EE113 pKa = 4.68TPWEE117 pKa = 3.87LVYY120 pKa = 10.68KK121 pKa = 10.83VEE123 pKa = 4.75DD124 pKa = 3.71SNVIDD129 pKa = 3.77GTTFAQIKK137 pKa = 10.06GKK139 pKa = 10.65LKK141 pKa = 10.61LSSAKK146 pKa = 10.11HH147 pKa = 4.86STDD150 pKa = 2.97IRR152 pKa = 11.84FKK154 pKa = 10.8PPTINILSKK163 pKa = 11.0DD164 pKa = 3.7FTKK167 pKa = 10.83DD168 pKa = 3.55CVDD171 pKa = 4.22FWSVDD176 pKa = 2.94KK177 pKa = 10.67PKK179 pKa = 10.5PIRR182 pKa = 11.84RR183 pKa = 11.84LLNPGPGYY191 pKa = 10.89GPDD194 pKa = 2.72GHH196 pKa = 6.38EE197 pKa = 3.62RR198 pKa = 11.84HH199 pKa = 6.03KK200 pKa = 10.82PIMLQPGEE208 pKa = 4.1TWATRR213 pKa = 11.84STIGRR218 pKa = 11.84TTSMRR223 pKa = 11.84YY224 pKa = 9.88APTEE228 pKa = 4.28RR229 pKa = 11.84IALDD233 pKa = 4.06DD234 pKa = 4.46KK235 pKa = 10.8PSCSEE240 pKa = 3.54AEE242 pKa = 4.11YY243 pKa = 10.08PLKK246 pKa = 10.78HH247 pKa = 5.04MHH249 pKa = 6.96KK250 pKa = 10.15LPEE253 pKa = 4.5SSLDD257 pKa = 3.61PGDD260 pKa = 4.45SVSQTTSNAMTKK272 pKa = 10.02QDD274 pKa = 3.42IEE276 pKa = 4.25EE277 pKa = 4.99LIEE280 pKa = 4.04TTINKK285 pKa = 9.49CLITQRR291 pKa = 11.84STISKK296 pKa = 9.61PLL298 pKa = 3.49
MM1 pKa = 8.04DD2 pKa = 5.02NYY4 pKa = 11.14SGAVVNNKK12 pKa = 9.82YY13 pKa = 10.02IEE15 pKa = 4.33SKK17 pKa = 9.13RR18 pKa = 11.84CEE20 pKa = 3.84YY21 pKa = 10.88RR22 pKa = 11.84LTNNEE27 pKa = 4.22TPIMLQFPSSLEE39 pKa = 3.87QTKK42 pKa = 10.0VRR44 pKa = 11.84MLGKK48 pKa = 10.07CMKK51 pKa = 9.46VDD53 pKa = 4.32HH54 pKa = 6.68IVIEE58 pKa = 4.22YY59 pKa = 10.2RR60 pKa = 11.84NQVPFNATGSVIVTIRR76 pKa = 11.84DD77 pKa = 3.21TRR79 pKa = 11.84LSHH82 pKa = 5.99EE83 pKa = 4.3QAAQAAFTFPIACNVDD99 pKa = 2.84LHH101 pKa = 6.36YY102 pKa = 10.7FSSSFFSLKK111 pKa = 10.84DD112 pKa = 3.25EE113 pKa = 4.68TPWEE117 pKa = 3.87LVYY120 pKa = 10.68KK121 pKa = 10.83VEE123 pKa = 4.75DD124 pKa = 3.71SNVIDD129 pKa = 3.77GTTFAQIKK137 pKa = 10.06GKK139 pKa = 10.65LKK141 pKa = 10.61LSSAKK146 pKa = 10.11HH147 pKa = 4.86STDD150 pKa = 2.97IRR152 pKa = 11.84FKK154 pKa = 10.8PPTINILSKK163 pKa = 11.0DD164 pKa = 3.7FTKK167 pKa = 10.83DD168 pKa = 3.55CVDD171 pKa = 4.22FWSVDD176 pKa = 2.94KK177 pKa = 10.67PKK179 pKa = 10.5PIRR182 pKa = 11.84RR183 pKa = 11.84LLNPGPGYY191 pKa = 10.89GPDD194 pKa = 2.72GHH196 pKa = 6.38EE197 pKa = 3.62RR198 pKa = 11.84HH199 pKa = 6.03KK200 pKa = 10.82PIMLQPGEE208 pKa = 4.1TWATRR213 pKa = 11.84STIGRR218 pKa = 11.84TTSMRR223 pKa = 11.84YY224 pKa = 9.88APTEE228 pKa = 4.28RR229 pKa = 11.84IALDD233 pKa = 4.06DD234 pKa = 4.46KK235 pKa = 10.8PSCSEE240 pKa = 3.54AEE242 pKa = 4.11YY243 pKa = 10.08PLKK246 pKa = 10.78HH247 pKa = 5.04MHH249 pKa = 6.96KK250 pKa = 10.15LPEE253 pKa = 4.5SSLDD257 pKa = 3.61PGDD260 pKa = 4.45SVSQTTSNAMTKK272 pKa = 10.02QDD274 pKa = 3.42IEE276 pKa = 4.25EE277 pKa = 4.99LIEE280 pKa = 4.04TTINKK285 pKa = 9.49CLITQRR291 pKa = 11.84STISKK296 pKa = 9.61PLL298 pKa = 3.49
Molecular weight: 33.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7TPF7|C7TPF7_9GEMI Replication enhancer OS=Rhynchosia yellow mosaic virus OX=529680 GN=ac3 PE=3 SV=1
MM1 pKa = 7.77VLILGCFLMVIHH13 pKa = 5.91YY14 pKa = 9.11VVVYY18 pKa = 7.8TIKK21 pKa = 10.54PINYY25 pKa = 8.58SLFLARR31 pKa = 11.84ILTTGYY37 pKa = 11.78SMMKK41 pKa = 10.17LAQNLITITQIVLHH55 pKa = 6.54RR56 pKa = 11.84SGTRR60 pKa = 11.84LIIIHH65 pKa = 5.99VKK67 pKa = 10.02NLSKK71 pKa = 10.16IHH73 pKa = 6.31RR74 pKa = 11.84RR75 pKa = 11.84SKK77 pKa = 10.41RR78 pKa = 11.84PPVSNEE84 pKa = 3.87LKK86 pKa = 9.08HH87 pKa = 5.66HH88 pKa = 5.16TVGVILGLDD97 pKa = 3.59VFIHH101 pKa = 6.67PNFPCNVNRR110 pKa = 11.84LDD112 pKa = 3.96AKK114 pKa = 10.07TFADD118 pKa = 3.69TMSNTVTTCHH128 pKa = 6.23IRR130 pKa = 11.84NAYY133 pKa = 9.83HH134 pKa = 6.26LTNMM138 pKa = 4.64
MM1 pKa = 7.77VLILGCFLMVIHH13 pKa = 5.91YY14 pKa = 9.11VVVYY18 pKa = 7.8TIKK21 pKa = 10.54PINYY25 pKa = 8.58SLFLARR31 pKa = 11.84ILTTGYY37 pKa = 11.78SMMKK41 pKa = 10.17LAQNLITITQIVLHH55 pKa = 6.54RR56 pKa = 11.84SGTRR60 pKa = 11.84LIIIHH65 pKa = 5.99VKK67 pKa = 10.02NLSKK71 pKa = 10.16IHH73 pKa = 6.31RR74 pKa = 11.84RR75 pKa = 11.84SKK77 pKa = 10.41RR78 pKa = 11.84PPVSNEE84 pKa = 3.87LKK86 pKa = 9.08HH87 pKa = 5.66HH88 pKa = 5.16TVGVILGLDD97 pKa = 3.59VFIHH101 pKa = 6.67PNFPCNVNRR110 pKa = 11.84LDD112 pKa = 3.96AKK114 pKa = 10.07TFADD118 pKa = 3.69TMSNTVTTCHH128 pKa = 6.23IRR130 pKa = 11.84NAYY133 pKa = 9.83HH134 pKa = 6.26LTNMM138 pKa = 4.64
Molecular weight: 15.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1803 |
97 |
364 |
200.3 |
22.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.713 ± 0.583 | 2.219 ± 0.306 |
4.603 ± 0.454 | 4.215 ± 0.637 |
4.382 ± 0.373 | 5.214 ± 0.572 |
3.938 ± 0.434 | 5.214 ± 0.754 |
6.434 ± 0.327 | 8.319 ± 0.858 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.662 ± 0.419 | 5.935 ± 0.556 |
5.99 ± 0.592 | 4.104 ± 0.457 |
6.6 ± 0.821 | 7.377 ± 0.621 |
6.045 ± 0.831 | 6.101 ± 0.677 |
0.998 ± 0.154 | 3.938 ± 0.418 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
