
Gaiella sp. SCGC AG-212-M14
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Rubrobacteria; Gaiellales; Gaiellaceae; Gaiella; unclassified Gaiella
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
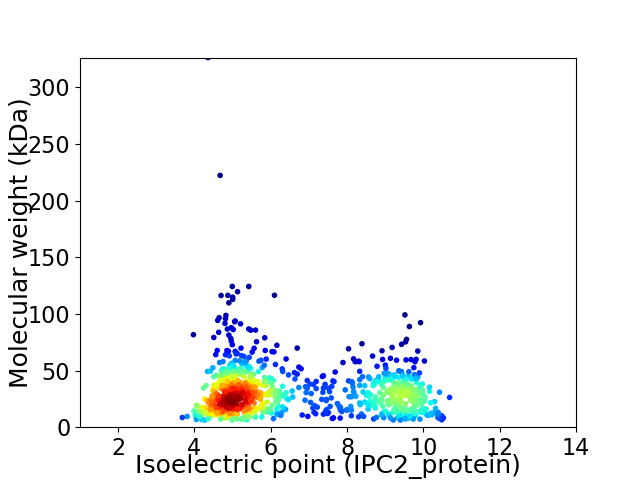
Virtual 2D-PAGE plot for 915 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177R6C3|A0A177R6C3_9ACTN Glyco_trans_2-like domain-containing protein OS=Gaiella sp. SCGC AG-212-M14 OX=1799656 GN=AYO48_03790 PE=4 SV=1
MM1 pKa = 8.19DD2 pKa = 3.64GRR4 pKa = 11.84AAAFQSVDD12 pKa = 3.02IEE14 pKa = 4.18IDD16 pKa = 2.8AGRR19 pKa = 11.84PLGIRR24 pKa = 11.84VAWAGGGAHH33 pKa = 6.92FLAIIGYY40 pKa = 9.68LEE42 pKa = 4.72DD43 pKa = 3.66VEE45 pKa = 5.65NYY47 pKa = 9.79IAVDD51 pKa = 3.42DD52 pKa = 4.96PIYY55 pKa = 10.85GKK57 pKa = 10.62SDD59 pKa = 3.24LTYY62 pKa = 8.73DD63 pKa = 3.9TLRR66 pKa = 11.84SSYY69 pKa = 10.16QGSGTWTHH77 pKa = 6.17TYY79 pKa = 8.19YY80 pKa = 10.21TQAA83 pKa = 3.88
MM1 pKa = 8.19DD2 pKa = 3.64GRR4 pKa = 11.84AAAFQSVDD12 pKa = 3.02IEE14 pKa = 4.18IDD16 pKa = 2.8AGRR19 pKa = 11.84PLGIRR24 pKa = 11.84VAWAGGGAHH33 pKa = 6.92FLAIIGYY40 pKa = 9.68LEE42 pKa = 4.72DD43 pKa = 3.66VEE45 pKa = 5.65NYY47 pKa = 9.79IAVDD51 pKa = 3.42DD52 pKa = 4.96PIYY55 pKa = 10.85GKK57 pKa = 10.62SDD59 pKa = 3.24LTYY62 pKa = 8.73DD63 pKa = 3.9TLRR66 pKa = 11.84SSYY69 pKa = 10.16QGSGTWTHH77 pKa = 6.17TYY79 pKa = 8.19YY80 pKa = 10.21TQAA83 pKa = 3.88
Molecular weight: 9.06 kDa
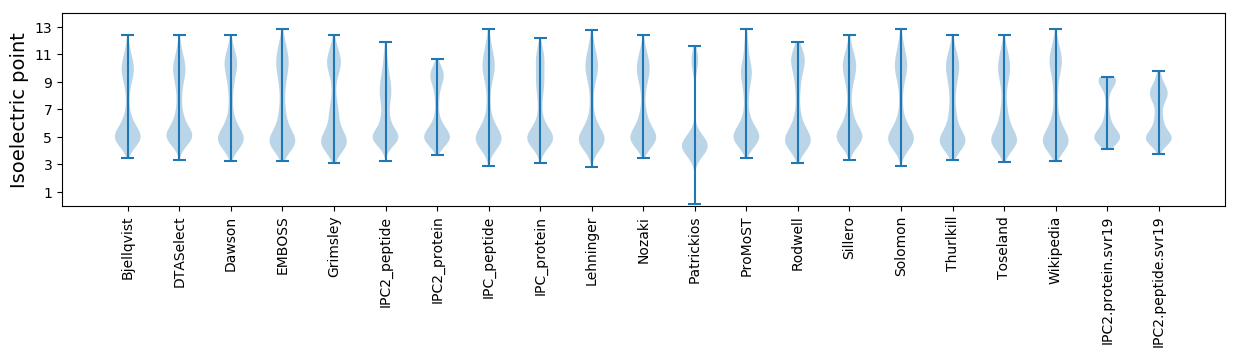
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177R532|A0A177R532_9ACTN Uncharacterized protein OS=Gaiella sp. SCGC AG-212-M14 OX=1799656 GN=AYO48_00780 PE=4 SV=1
MM1 pKa = 7.47SPASTEE7 pKa = 3.8QRR9 pKa = 11.84SRR11 pKa = 11.84ASIRR15 pKa = 11.84RR16 pKa = 11.84LIAPLALAQFICSFAGSNMNVMISDD41 pKa = 3.71ISKK44 pKa = 10.76DD45 pKa = 3.3LNTTVKK51 pKa = 10.42GVQTTITLFLLIMAILMIPCSKK73 pKa = 9.91LTDD76 pKa = 2.64RR77 pKa = 11.84WGRR80 pKa = 11.84KK81 pKa = 8.17RR82 pKa = 11.84CFTMGLTLYY91 pKa = 10.93GIGALVSAAAPNLGVLILGNSVFEE115 pKa = 4.51GVGTALLIPPVYY127 pKa = 10.28ILATLWFADD136 pKa = 3.54VTSRR140 pKa = 11.84AWAFGVISGLGGVGAAAGPLIGGVITTGISWRR172 pKa = 11.84AAFVFQALVIATILLLSRR190 pKa = 11.84RR191 pKa = 11.84LVDD194 pKa = 4.82PLPADD199 pKa = 3.55PARR202 pKa = 11.84PFDD205 pKa = 3.84AVGAILSAVGMFFVVFGILQAGSNNVLFVICLGIGAAFLASFFLFIRR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84EE255 pKa = 4.08RR256 pKa = 11.84AGKK259 pKa = 10.03EE260 pKa = 3.67PLLSTRR266 pKa = 11.84LFRR269 pKa = 11.84NRR271 pKa = 11.84TCNLALVTQNVQWLMLLGTSFVVSVFLQTEE301 pKa = 4.41RR302 pKa = 11.84GYY304 pKa = 11.04SAIKK308 pKa = 9.21TGVIFTAATVGILISSFAAEE328 pKa = 3.67RR329 pKa = 11.84WARR332 pKa = 11.84KK333 pKa = 8.75YY334 pKa = 10.08AQKK337 pKa = 9.72TLIVTGFVVTIVGMGLLLGLVKK359 pKa = 10.71ASSSDD364 pKa = 3.02WAFVPGLFLIGFGVGAMLTPSVNVVQSSFPEE395 pKa = 3.99ALQGEE400 pKa = 4.57ISGLSRR406 pKa = 11.84SVSNLGSSFGTAIAGTIIVSVVAPGYY432 pKa = 10.2RR433 pKa = 11.84SYY435 pKa = 11.71AWALVSLVLIGLIGLGAALLLPSKK459 pKa = 10.65LEE461 pKa = 4.07RR462 pKa = 11.84PAA464 pKa = 5.03
MM1 pKa = 7.47SPASTEE7 pKa = 3.8QRR9 pKa = 11.84SRR11 pKa = 11.84ASIRR15 pKa = 11.84RR16 pKa = 11.84LIAPLALAQFICSFAGSNMNVMISDD41 pKa = 3.71ISKK44 pKa = 10.76DD45 pKa = 3.3LNTTVKK51 pKa = 10.42GVQTTITLFLLIMAILMIPCSKK73 pKa = 9.91LTDD76 pKa = 2.64RR77 pKa = 11.84WGRR80 pKa = 11.84KK81 pKa = 8.17RR82 pKa = 11.84CFTMGLTLYY91 pKa = 10.93GIGALVSAAAPNLGVLILGNSVFEE115 pKa = 4.51GVGTALLIPPVYY127 pKa = 10.28ILATLWFADD136 pKa = 3.54VTSRR140 pKa = 11.84AWAFGVISGLGGVGAAAGPLIGGVITTGISWRR172 pKa = 11.84AAFVFQALVIATILLLSRR190 pKa = 11.84RR191 pKa = 11.84LVDD194 pKa = 4.82PLPADD199 pKa = 3.55PARR202 pKa = 11.84PFDD205 pKa = 3.84AVGAILSAVGMFFVVFGILQAGSNNVLFVICLGIGAAFLASFFLFIRR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84EE255 pKa = 4.08RR256 pKa = 11.84AGKK259 pKa = 10.03EE260 pKa = 3.67PLLSTRR266 pKa = 11.84LFRR269 pKa = 11.84NRR271 pKa = 11.84TCNLALVTQNVQWLMLLGTSFVVSVFLQTEE301 pKa = 4.41RR302 pKa = 11.84GYY304 pKa = 11.04SAIKK308 pKa = 9.21TGVIFTAATVGILISSFAAEE328 pKa = 3.67RR329 pKa = 11.84WARR332 pKa = 11.84KK333 pKa = 8.75YY334 pKa = 10.08AQKK337 pKa = 9.72TLIVTGFVVTIVGMGLLLGLVKK359 pKa = 10.71ASSSDD364 pKa = 3.02WAFVPGLFLIGFGVGAMLTPSVNVVQSSFPEE395 pKa = 3.99ALQGEE400 pKa = 4.57ISGLSRR406 pKa = 11.84SVSNLGSSFGTAIAGTIIVSVVAPGYY432 pKa = 10.2RR433 pKa = 11.84SYY435 pKa = 11.71AWALVSLVLIGLIGLGAALLLPSKK459 pKa = 10.65LEE461 pKa = 4.07RR462 pKa = 11.84PAA464 pKa = 5.03
Molecular weight: 48.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
275724 |
53 |
3217 |
301.3 |
32.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.604 ± 0.11 | 0.733 ± 0.021 |
5.461 ± 0.059 | 6.116 ± 0.126 |
3.493 ± 0.048 | 8.898 ± 0.074 |
2.034 ± 0.042 | 4.292 ± 0.059 |
2.489 ± 0.068 | 10.751 ± 0.128 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.591 ± 0.033 | 2.189 ± 0.061 |
5.307 ± 0.054 | 2.865 ± 0.046 |
7.798 ± 0.115 | 5.646 ± 0.118 |
5.507 ± 0.138 | 8.502 ± 0.076 |
1.534 ± 0.037 | 2.191 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
