
Batrachochytrium dendrobatidis (strain JAM81 / FGSC 10211) (Frog chytrid fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Chytridiomycota; Chytridiomycota incertae sedis; Chytridiomycetes; Rhizophydiales; Rhizophydiales incertae sedis; Batrachochytrium; Batrachochytrium dendrobatidis
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
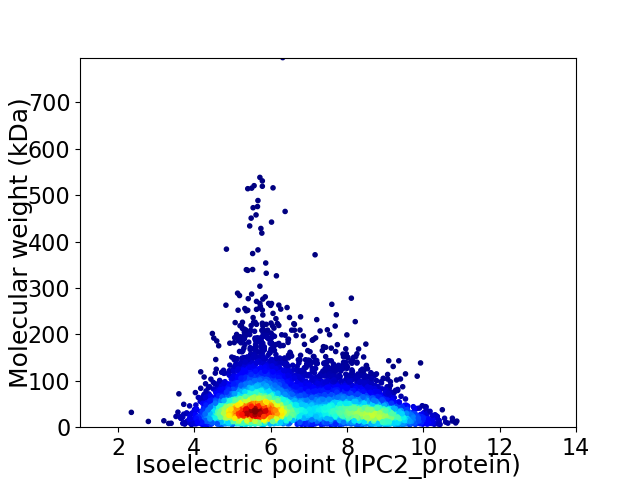
Virtual 2D-PAGE plot for 8610 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F4PF81|F4PF81_BATDJ Sulfhydryl oxidase OS=Batrachochytrium dendrobatidis (strain JAM81 / FGSC 10211) OX=684364 GN=BATDEDRAFT_15048 PE=4 SV=1
MM1 pKa = 7.62HH2 pKa = 7.64LPLQLQPYY10 pKa = 9.57ADD12 pKa = 3.62VLAEE16 pKa = 4.1TKK18 pKa = 10.69KK19 pKa = 10.56PGFIINGKK27 pKa = 8.9PEE29 pKa = 4.29STTPLQSKK37 pKa = 10.11FSGRR41 pKa = 11.84PYY43 pKa = 9.66WLKK46 pKa = 11.03ADD48 pKa = 5.05AYY50 pKa = 9.74PITQNGQPLRR60 pKa = 11.84LLAQINFSEE69 pKa = 4.38MEE71 pKa = 4.09EE72 pKa = 4.31TIPDD76 pKa = 3.88YY77 pKa = 9.13PTEE80 pKa = 4.95GILQFFVADD89 pKa = 3.85DD90 pKa = 3.91DD91 pKa = 5.32VYY93 pKa = 11.71GLNFDD98 pKa = 5.46DD99 pKa = 5.47GTNQDD104 pKa = 3.49TFRR107 pKa = 11.84VVYY110 pKa = 10.35HH111 pKa = 5.93EE112 pKa = 4.25TVEE115 pKa = 4.06MDD117 pKa = 3.5PSKK120 pKa = 10.89WMTDD124 pKa = 3.55FPEE127 pKa = 4.74FGNEE131 pKa = 3.48NYY133 pKa = 10.1FPVEE137 pKa = 4.1KK138 pKa = 10.16EE139 pKa = 3.9CAISFDD145 pKa = 3.56VSEE148 pKa = 5.45EE149 pKa = 3.96IMSSSDD155 pKa = 3.01YY156 pKa = 10.45RR157 pKa = 11.84FNIITEE163 pKa = 4.02VDD165 pKa = 3.29QLLEE169 pKa = 4.36SGTDD173 pKa = 3.1EE174 pKa = 5.81DD175 pKa = 4.8YY176 pKa = 11.25AAHH179 pKa = 7.08DD180 pKa = 4.91EE181 pKa = 4.69IMDD184 pKa = 4.6AYY186 pKa = 10.8HH187 pKa = 7.23EE188 pKa = 4.33IASGQGCKK196 pKa = 10.34LGGYY200 pKa = 9.26SFFTQEE206 pKa = 4.29DD207 pKa = 3.42PRR209 pKa = 11.84AYY211 pKa = 10.38GDD213 pKa = 3.99YY214 pKa = 9.55LTHH217 pKa = 7.22DD218 pKa = 4.04TLLLQIDD225 pKa = 4.56SDD227 pKa = 4.4DD228 pKa = 4.84DD229 pKa = 3.97MNIMWGDD236 pKa = 3.31TGVANFFISSEE247 pKa = 3.99DD248 pKa = 3.28LKK250 pKa = 11.48NRR252 pKa = 11.84DD253 pKa = 3.39FSKK256 pKa = 11.02VLYY259 pKa = 10.64NWDD262 pKa = 3.64CYY264 pKa = 11.2
MM1 pKa = 7.62HH2 pKa = 7.64LPLQLQPYY10 pKa = 9.57ADD12 pKa = 3.62VLAEE16 pKa = 4.1TKK18 pKa = 10.69KK19 pKa = 10.56PGFIINGKK27 pKa = 8.9PEE29 pKa = 4.29STTPLQSKK37 pKa = 10.11FSGRR41 pKa = 11.84PYY43 pKa = 9.66WLKK46 pKa = 11.03ADD48 pKa = 5.05AYY50 pKa = 9.74PITQNGQPLRR60 pKa = 11.84LLAQINFSEE69 pKa = 4.38MEE71 pKa = 4.09EE72 pKa = 4.31TIPDD76 pKa = 3.88YY77 pKa = 9.13PTEE80 pKa = 4.95GILQFFVADD89 pKa = 3.85DD90 pKa = 3.91DD91 pKa = 5.32VYY93 pKa = 11.71GLNFDD98 pKa = 5.46DD99 pKa = 5.47GTNQDD104 pKa = 3.49TFRR107 pKa = 11.84VVYY110 pKa = 10.35HH111 pKa = 5.93EE112 pKa = 4.25TVEE115 pKa = 4.06MDD117 pKa = 3.5PSKK120 pKa = 10.89WMTDD124 pKa = 3.55FPEE127 pKa = 4.74FGNEE131 pKa = 3.48NYY133 pKa = 10.1FPVEE137 pKa = 4.1KK138 pKa = 10.16EE139 pKa = 3.9CAISFDD145 pKa = 3.56VSEE148 pKa = 5.45EE149 pKa = 3.96IMSSSDD155 pKa = 3.01YY156 pKa = 10.45RR157 pKa = 11.84FNIITEE163 pKa = 4.02VDD165 pKa = 3.29QLLEE169 pKa = 4.36SGTDD173 pKa = 3.1EE174 pKa = 5.81DD175 pKa = 4.8YY176 pKa = 11.25AAHH179 pKa = 7.08DD180 pKa = 4.91EE181 pKa = 4.69IMDD184 pKa = 4.6AYY186 pKa = 10.8HH187 pKa = 7.23EE188 pKa = 4.33IASGQGCKK196 pKa = 10.34LGGYY200 pKa = 9.26SFFTQEE206 pKa = 4.29DD207 pKa = 3.42PRR209 pKa = 11.84AYY211 pKa = 10.38GDD213 pKa = 3.99YY214 pKa = 9.55LTHH217 pKa = 7.22DD218 pKa = 4.04TLLLQIDD225 pKa = 4.56SDD227 pKa = 4.4DD228 pKa = 4.84DD229 pKa = 3.97MNIMWGDD236 pKa = 3.31TGVANFFISSEE247 pKa = 3.99DD248 pKa = 3.28LKK250 pKa = 11.48NRR252 pKa = 11.84DD253 pKa = 3.39FSKK256 pKa = 11.02VLYY259 pKa = 10.64NWDD262 pKa = 3.64CYY264 pKa = 11.2
Molecular weight: 30.28 kDa
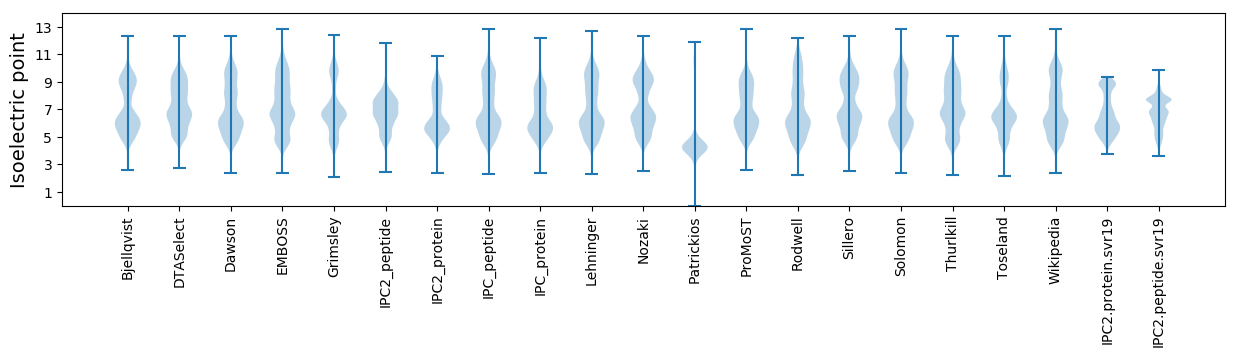
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F4P5P6|F4P5P6_BATDJ 40S ribosomal protein S4 OS=Batrachochytrium dendrobatidis (strain JAM81 / FGSC 10211) OX=684364 GN=BATDEDRAFT_35382 PE=3 SV=1
MM1 pKa = 8.06DD2 pKa = 4.77SADD5 pKa = 3.88TACSSVKK12 pKa = 10.0ICLVPLAALNLSKK25 pKa = 10.85QITEE29 pKa = 3.97EE30 pKa = 4.1LLCQFFSFIGPIVSISLQPCEE51 pKa = 4.73ANPLVQEE58 pKa = 4.57SLVQFQDD65 pKa = 3.72SEE67 pKa = 4.37DD68 pKa = 3.58AALALHH74 pKa = 6.18LTGTVLADD82 pKa = 3.22KK83 pKa = 10.68ALFITPPSAKK93 pKa = 8.8IQSFNSHH100 pKa = 6.41GYY102 pKa = 10.85LNIPPPNPTAPGVLVPGSMAGSTIVPYY129 pKa = 10.71GQTATIAGQDD139 pKa = 3.53SVDD142 pKa = 3.28RR143 pKa = 11.84TIYY146 pKa = 9.5TGNIHH151 pKa = 6.82SGLSQQEE158 pKa = 3.86VSMLFSSCGDD168 pKa = 3.29VTQVKK173 pKa = 8.66MAGDD177 pKa = 3.83ATHH180 pKa = 5.59STRR183 pKa = 11.84YY184 pKa = 10.43AFIEE188 pKa = 4.35FATSEE193 pKa = 4.34SAAMALNLHH202 pKa = 5.72GMMVAGRR209 pKa = 11.84AIKK212 pKa = 10.44VNRR215 pKa = 11.84SKK217 pKa = 11.07HH218 pKa = 5.64SIGRR222 pKa = 11.84PIGMYY227 pKa = 9.95PGMFRR232 pKa = 11.84PDD234 pKa = 2.78TDD236 pKa = 3.25MVMKK240 pKa = 10.35QAMQAQMRR248 pKa = 11.84MGHH251 pKa = 5.8SFSGDD256 pKa = 2.81ARR258 pKa = 11.84SAGKK262 pKa = 10.03FPIIPGTPGMMAIAAAANVPPGTNVMPPMTSYY294 pKa = 11.05LSAMHH299 pKa = 6.99APNPLANINAAATSTAARR317 pKa = 11.84TSQEE321 pKa = 3.5KK322 pKa = 10.09SSRR325 pKa = 11.84GRR327 pKa = 11.84TFSRR331 pKa = 11.84EE332 pKa = 3.7RR333 pKa = 11.84GRR335 pKa = 11.84DD336 pKa = 3.05RR337 pKa = 11.84HH338 pKa = 5.58EE339 pKa = 4.91SISPEE344 pKa = 3.67SSSRR348 pKa = 11.84RR349 pKa = 11.84KK350 pKa = 8.54SARR353 pKa = 11.84RR354 pKa = 11.84RR355 pKa = 11.84SYY357 pKa = 10.49SRR359 pKa = 11.84SPATSRR365 pKa = 11.84SRR367 pKa = 11.84SVSPRR372 pKa = 11.84PKK374 pKa = 9.91DD375 pKa = 3.31SRR377 pKa = 11.84RR378 pKa = 11.84AKK380 pKa = 10.5DD381 pKa = 3.16KK382 pKa = 11.43SRR384 pKa = 11.84DD385 pKa = 3.28RR386 pKa = 11.84SHH388 pKa = 8.44AMDD391 pKa = 4.47RR392 pKa = 11.84PRR394 pKa = 11.84QRR396 pKa = 11.84DD397 pKa = 3.0RR398 pKa = 11.84SRR400 pKa = 11.84QRR402 pKa = 11.84DD403 pKa = 3.13RR404 pKa = 11.84SRR406 pKa = 11.84DD407 pKa = 3.19HH408 pKa = 6.1GRR410 pKa = 11.84NRR412 pKa = 11.84SRR414 pKa = 11.84LRR416 pKa = 11.84SRR418 pKa = 11.84DD419 pKa = 3.01RR420 pKa = 11.84DD421 pKa = 3.34RR422 pKa = 11.84SRR424 pKa = 11.84DD425 pKa = 3.45RR426 pKa = 11.84ASGRR430 pKa = 11.84DD431 pKa = 3.24RR432 pKa = 11.84YY433 pKa = 10.14RR434 pKa = 11.84EE435 pKa = 3.68RR436 pKa = 11.84DD437 pKa = 3.12RR438 pKa = 11.84DD439 pKa = 3.34MDD441 pKa = 3.8RR442 pKa = 11.84SKK444 pKa = 11.34DD445 pKa = 3.42RR446 pKa = 11.84SRR448 pKa = 11.84DD449 pKa = 3.07RR450 pKa = 11.84SRR452 pKa = 11.84DD453 pKa = 3.19RR454 pKa = 11.84RR455 pKa = 11.84VEE457 pKa = 3.68QSSRR461 pKa = 11.84NRR463 pKa = 11.84NRR465 pKa = 11.84TRR467 pKa = 11.84NSSTSRR473 pKa = 11.84RR474 pKa = 11.84VRR476 pKa = 11.84GDD478 pKa = 2.86SRR480 pKa = 11.84SRR482 pKa = 11.84SLAAGAKK489 pKa = 9.95AGGSFGDD496 pKa = 4.34LRR498 pKa = 11.84AILNAKK504 pKa = 9.65HH505 pKa = 6.87DD506 pKa = 3.85GDD508 pKa = 4.26KK509 pKa = 10.92RR510 pKa = 11.84SGSQRR515 pKa = 11.84TSQDD519 pKa = 2.8HH520 pKa = 5.79GVVSVSGEE528 pKa = 4.2HH529 pKa = 6.51EE530 pKa = 4.79DD531 pKa = 5.27KK532 pKa = 10.72PDD534 pKa = 3.34APIPSKK540 pKa = 11.18DD541 pKa = 4.38LDD543 pKa = 3.38TDD545 pKa = 3.71MQIDD549 pKa = 3.73SGDD552 pKa = 3.47YY553 pKa = 10.2SAA555 pKa = 5.77
MM1 pKa = 8.06DD2 pKa = 4.77SADD5 pKa = 3.88TACSSVKK12 pKa = 10.0ICLVPLAALNLSKK25 pKa = 10.85QITEE29 pKa = 3.97EE30 pKa = 4.1LLCQFFSFIGPIVSISLQPCEE51 pKa = 4.73ANPLVQEE58 pKa = 4.57SLVQFQDD65 pKa = 3.72SEE67 pKa = 4.37DD68 pKa = 3.58AALALHH74 pKa = 6.18LTGTVLADD82 pKa = 3.22KK83 pKa = 10.68ALFITPPSAKK93 pKa = 8.8IQSFNSHH100 pKa = 6.41GYY102 pKa = 10.85LNIPPPNPTAPGVLVPGSMAGSTIVPYY129 pKa = 10.71GQTATIAGQDD139 pKa = 3.53SVDD142 pKa = 3.28RR143 pKa = 11.84TIYY146 pKa = 9.5TGNIHH151 pKa = 6.82SGLSQQEE158 pKa = 3.86VSMLFSSCGDD168 pKa = 3.29VTQVKK173 pKa = 8.66MAGDD177 pKa = 3.83ATHH180 pKa = 5.59STRR183 pKa = 11.84YY184 pKa = 10.43AFIEE188 pKa = 4.35FATSEE193 pKa = 4.34SAAMALNLHH202 pKa = 5.72GMMVAGRR209 pKa = 11.84AIKK212 pKa = 10.44VNRR215 pKa = 11.84SKK217 pKa = 11.07HH218 pKa = 5.64SIGRR222 pKa = 11.84PIGMYY227 pKa = 9.95PGMFRR232 pKa = 11.84PDD234 pKa = 2.78TDD236 pKa = 3.25MVMKK240 pKa = 10.35QAMQAQMRR248 pKa = 11.84MGHH251 pKa = 5.8SFSGDD256 pKa = 2.81ARR258 pKa = 11.84SAGKK262 pKa = 10.03FPIIPGTPGMMAIAAAANVPPGTNVMPPMTSYY294 pKa = 11.05LSAMHH299 pKa = 6.99APNPLANINAAATSTAARR317 pKa = 11.84TSQEE321 pKa = 3.5KK322 pKa = 10.09SSRR325 pKa = 11.84GRR327 pKa = 11.84TFSRR331 pKa = 11.84EE332 pKa = 3.7RR333 pKa = 11.84GRR335 pKa = 11.84DD336 pKa = 3.05RR337 pKa = 11.84HH338 pKa = 5.58EE339 pKa = 4.91SISPEE344 pKa = 3.67SSSRR348 pKa = 11.84RR349 pKa = 11.84KK350 pKa = 8.54SARR353 pKa = 11.84RR354 pKa = 11.84RR355 pKa = 11.84SYY357 pKa = 10.49SRR359 pKa = 11.84SPATSRR365 pKa = 11.84SRR367 pKa = 11.84SVSPRR372 pKa = 11.84PKK374 pKa = 9.91DD375 pKa = 3.31SRR377 pKa = 11.84RR378 pKa = 11.84AKK380 pKa = 10.5DD381 pKa = 3.16KK382 pKa = 11.43SRR384 pKa = 11.84DD385 pKa = 3.28RR386 pKa = 11.84SHH388 pKa = 8.44AMDD391 pKa = 4.47RR392 pKa = 11.84PRR394 pKa = 11.84QRR396 pKa = 11.84DD397 pKa = 3.0RR398 pKa = 11.84SRR400 pKa = 11.84QRR402 pKa = 11.84DD403 pKa = 3.13RR404 pKa = 11.84SRR406 pKa = 11.84DD407 pKa = 3.19HH408 pKa = 6.1GRR410 pKa = 11.84NRR412 pKa = 11.84SRR414 pKa = 11.84LRR416 pKa = 11.84SRR418 pKa = 11.84DD419 pKa = 3.01RR420 pKa = 11.84DD421 pKa = 3.34RR422 pKa = 11.84SRR424 pKa = 11.84DD425 pKa = 3.45RR426 pKa = 11.84ASGRR430 pKa = 11.84DD431 pKa = 3.24RR432 pKa = 11.84YY433 pKa = 10.14RR434 pKa = 11.84EE435 pKa = 3.68RR436 pKa = 11.84DD437 pKa = 3.12RR438 pKa = 11.84DD439 pKa = 3.34MDD441 pKa = 3.8RR442 pKa = 11.84SKK444 pKa = 11.34DD445 pKa = 3.42RR446 pKa = 11.84SRR448 pKa = 11.84DD449 pKa = 3.07RR450 pKa = 11.84SRR452 pKa = 11.84DD453 pKa = 3.19RR454 pKa = 11.84RR455 pKa = 11.84VEE457 pKa = 3.68QSSRR461 pKa = 11.84NRR463 pKa = 11.84NRR465 pKa = 11.84TRR467 pKa = 11.84NSSTSRR473 pKa = 11.84RR474 pKa = 11.84VRR476 pKa = 11.84GDD478 pKa = 2.86SRR480 pKa = 11.84SRR482 pKa = 11.84SLAAGAKK489 pKa = 9.95AGGSFGDD496 pKa = 4.34LRR498 pKa = 11.84AILNAKK504 pKa = 9.65HH505 pKa = 6.87DD506 pKa = 3.85GDD508 pKa = 4.26KK509 pKa = 10.92RR510 pKa = 11.84SGSQRR515 pKa = 11.84TSQDD519 pKa = 2.8HH520 pKa = 5.79GVVSVSGEE528 pKa = 4.2HH529 pKa = 6.51EE530 pKa = 4.79DD531 pKa = 5.27KK532 pKa = 10.72PDD534 pKa = 3.34APIPSKK540 pKa = 11.18DD541 pKa = 4.38LDD543 pKa = 3.38TDD545 pKa = 3.71MQIDD549 pKa = 3.73SGDD552 pKa = 3.47YY553 pKa = 10.2SAA555 pKa = 5.77
Molecular weight: 60.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3887913 |
49 |
6976 |
451.6 |
50.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.991 ± 0.026 | 1.478 ± 0.012 |
5.671 ± 0.017 | 5.406 ± 0.024 |
3.941 ± 0.016 | 5.26 ± 0.024 |
2.694 ± 0.012 | 6.289 ± 0.018 |
5.727 ± 0.025 | 9.341 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.453 ± 0.01 | 4.551 ± 0.016 |
4.919 ± 0.021 | 4.592 ± 0.02 |
4.726 ± 0.016 | 9.424 ± 0.036 |
6.456 ± 0.019 | 6.107 ± 0.021 |
1.055 ± 0.008 | 2.918 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
