
Fomitopsis pinicola (strain FP-58527) (Brown rot fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Fomitopsidaceae; Fomitopsis; Fomitopsis pinicola
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
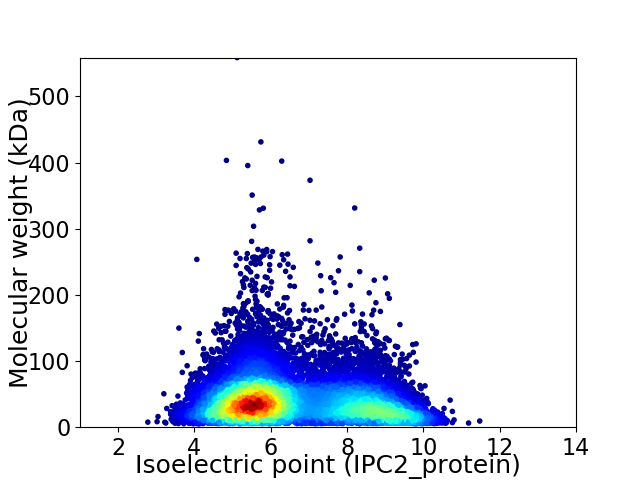
Virtual 2D-PAGE plot for 13806 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
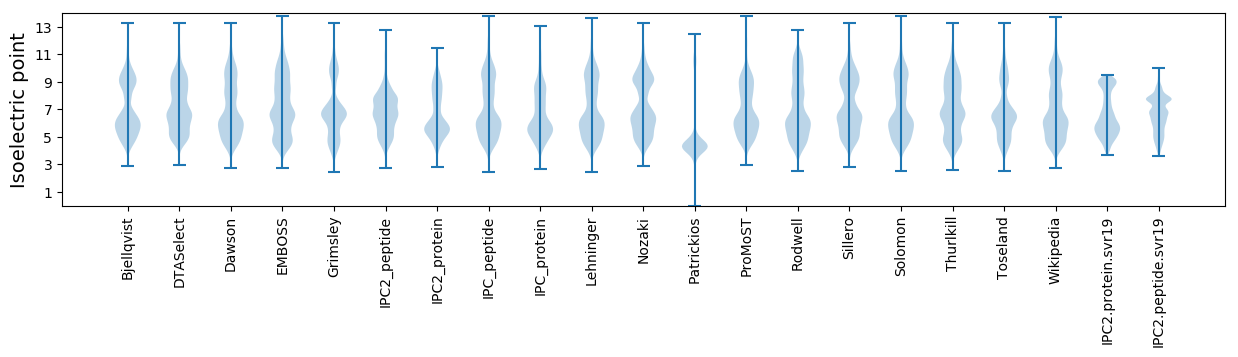
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S8EF35|S8EF35_FOMPI Uncharacterized protein OS=Fomitopsis pinicola (strain FP-58527) OX=743788 GN=FOMPIDRAFT_1115832 PE=4 SV=1
MM1 pKa = 7.45HH2 pKa = 7.27SAHH5 pKa = 6.62NLLALVTLAVLASAAPSVTDD25 pKa = 3.19TAKK28 pKa = 10.74RR29 pKa = 11.84STCTVDD35 pKa = 3.41SVSSASSLSDD45 pKa = 3.2CSDD48 pKa = 3.44VVISSFTVDD57 pKa = 4.63DD58 pKa = 4.5GDD60 pKa = 4.0TVTMSFASGATVTMTGEE77 pKa = 4.05VTFASTTASGPLITFEE93 pKa = 4.51GDD95 pKa = 3.38SISFDD100 pKa = 3.26GGGNTFNGNGADD112 pKa = 3.58YY113 pKa = 10.59WDD115 pKa = 4.3GEE117 pKa = 4.44GTNGGVAKK125 pKa = 8.72PHH127 pKa = 6.79PFMKK131 pKa = 10.13IKK133 pKa = 10.83ASGTFEE139 pKa = 3.95NFIVLNTPAQAISVGNDD156 pKa = 3.14DD157 pKa = 5.23PLTISDD163 pKa = 3.65VTVNNLAGNTDD174 pKa = 3.8SLGHH178 pKa = 5.69NTDD181 pKa = 3.41GFDD184 pKa = 3.3VAASDD189 pKa = 3.88VTIKK193 pKa = 11.01NCVVEE198 pKa = 4.32NQDD201 pKa = 3.22DD202 pKa = 4.37CIAINSGSNIVFQDD216 pKa = 3.55NTCSGGHH223 pKa = 5.65GMSIGSIASGKK234 pKa = 7.22TVSGVIFSGNTVSDD248 pKa = 3.6SLYY251 pKa = 7.96GTRR254 pKa = 11.84VKK256 pKa = 10.62VDD258 pKa = 3.15SDD260 pKa = 3.61ATSGSVSNVTWSGNTISGISDD281 pKa = 3.53YY282 pKa = 11.36GVLITQSYY290 pKa = 8.96PDD292 pKa = 3.61NDD294 pKa = 3.98GTPGTDD300 pKa = 3.12TTISDD305 pKa = 4.38LNFIDD310 pKa = 3.7STTSVSVDD318 pKa = 3.02SGATRR323 pKa = 11.84VTVDD327 pKa = 3.64CGYY330 pKa = 8.94CTGTWDD336 pKa = 3.99WSQLDD341 pKa = 3.81ATGGDD346 pKa = 3.22AGTIVASDD354 pKa = 3.35ATIEE358 pKa = 4.34GGTYY362 pKa = 10.41
MM1 pKa = 7.45HH2 pKa = 7.27SAHH5 pKa = 6.62NLLALVTLAVLASAAPSVTDD25 pKa = 3.19TAKK28 pKa = 10.74RR29 pKa = 11.84STCTVDD35 pKa = 3.41SVSSASSLSDD45 pKa = 3.2CSDD48 pKa = 3.44VVISSFTVDD57 pKa = 4.63DD58 pKa = 4.5GDD60 pKa = 4.0TVTMSFASGATVTMTGEE77 pKa = 4.05VTFASTTASGPLITFEE93 pKa = 4.51GDD95 pKa = 3.38SISFDD100 pKa = 3.26GGGNTFNGNGADD112 pKa = 3.58YY113 pKa = 10.59WDD115 pKa = 4.3GEE117 pKa = 4.44GTNGGVAKK125 pKa = 8.72PHH127 pKa = 6.79PFMKK131 pKa = 10.13IKK133 pKa = 10.83ASGTFEE139 pKa = 3.95NFIVLNTPAQAISVGNDD156 pKa = 3.14DD157 pKa = 5.23PLTISDD163 pKa = 3.65VTVNNLAGNTDD174 pKa = 3.8SLGHH178 pKa = 5.69NTDD181 pKa = 3.41GFDD184 pKa = 3.3VAASDD189 pKa = 3.88VTIKK193 pKa = 11.01NCVVEE198 pKa = 4.32NQDD201 pKa = 3.22DD202 pKa = 4.37CIAINSGSNIVFQDD216 pKa = 3.55NTCSGGHH223 pKa = 5.65GMSIGSIASGKK234 pKa = 7.22TVSGVIFSGNTVSDD248 pKa = 3.6SLYY251 pKa = 7.96GTRR254 pKa = 11.84VKK256 pKa = 10.62VDD258 pKa = 3.15SDD260 pKa = 3.61ATSGSVSNVTWSGNTISGISDD281 pKa = 3.53YY282 pKa = 11.36GVLITQSYY290 pKa = 8.96PDD292 pKa = 3.61NDD294 pKa = 3.98GTPGTDD300 pKa = 3.12TTISDD305 pKa = 4.38LNFIDD310 pKa = 3.7STTSVSVDD318 pKa = 3.02SGATRR323 pKa = 11.84VTVDD327 pKa = 3.64CGYY330 pKa = 8.94CTGTWDD336 pKa = 3.99WSQLDD341 pKa = 3.81ATGGDD346 pKa = 3.22AGTIVASDD354 pKa = 3.35ATIEE358 pKa = 4.34GGTYY362 pKa = 10.41
Molecular weight: 36.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S8FTB7|S8FTB7_FOMPI TPR_REGION domain-containing protein OS=Fomitopsis pinicola (strain FP-58527) OX=743788 GN=FOMPIDRAFT_1120590 PE=4 SV=1
MM1 pKa = 7.46RR2 pKa = 11.84PSWSGRR8 pKa = 11.84RR9 pKa = 11.84PPPVHH14 pKa = 6.58RR15 pKa = 11.84HH16 pKa = 5.1AAPPPTQAVAPRR28 pKa = 11.84SPPRR32 pKa = 11.84PPPIAPAPALAARR45 pKa = 11.84ALTLAARR52 pKa = 11.84LRR54 pKa = 11.84PRR56 pKa = 11.84LLPP59 pKa = 4.12
MM1 pKa = 7.46RR2 pKa = 11.84PSWSGRR8 pKa = 11.84RR9 pKa = 11.84PPPVHH14 pKa = 6.58RR15 pKa = 11.84HH16 pKa = 5.1AAPPPTQAVAPRR28 pKa = 11.84SPPRR32 pKa = 11.84PPPIAPAPALAARR45 pKa = 11.84ALTLAARR52 pKa = 11.84LRR54 pKa = 11.84PRR56 pKa = 11.84LLPP59 pKa = 4.12
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5596684 |
49 |
5042 |
405.4 |
44.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.521 ± 0.02 | 1.288 ± 0.01 |
5.712 ± 0.015 | 6.034 ± 0.021 |
3.459 ± 0.013 | 6.633 ± 0.019 |
2.617 ± 0.01 | 4.388 ± 0.015 |
4.168 ± 0.017 | 9.216 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.111 ± 0.008 | 3.034 ± 0.012 |
6.602 ± 0.027 | 3.676 ± 0.012 |
6.678 ± 0.021 | 8.051 ± 0.027 |
5.967 ± 0.015 | 6.633 ± 0.016 |
1.501 ± 0.006 | 2.71 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
