
Haloterrigena sp. YPL8
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Haloterrigena; unclassified Haloterrigena
Average proteome isoelectric point is 4.85
Get precalculated fractions of proteins
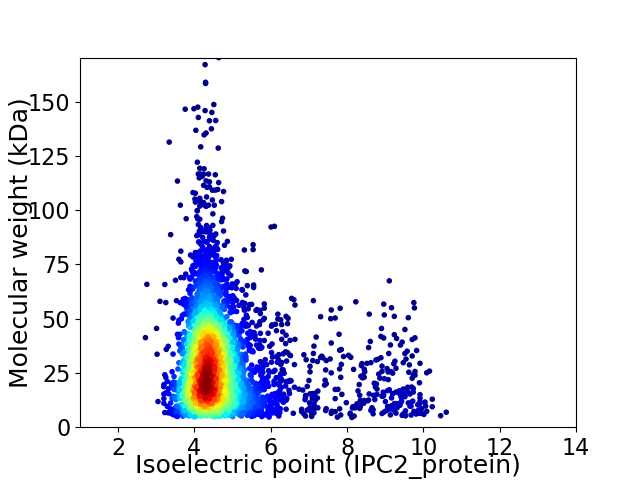
Virtual 2D-PAGE plot for 3981 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7D5GNG8|A0A7D5GNG8_9EURY Uncharacterized protein OS=Haloterrigena sp. YPL8 OX=1699371 GN=HYG82_11000 PE=4 SV=1
MM1 pKa = 6.59VTTGAWGGFHH11 pKa = 7.67RR12 pKa = 11.84MGCITRR18 pKa = 11.84RR19 pKa = 11.84DD20 pKa = 3.48ILKK23 pKa = 10.38ASLGFGTVMGVGLVGRR39 pKa = 11.84WPEE42 pKa = 4.1TTTSEE47 pKa = 4.19TSFGDD52 pKa = 3.83GVNLQPSYY60 pKa = 10.89FCSANQDD67 pKa = 3.39IGWDD71 pKa = 3.85FMSQYY76 pKa = 11.42SDD78 pKa = 2.75IQTLRR83 pKa = 11.84MRR85 pKa = 11.84IEE87 pKa = 4.06PFSFSEE93 pKa = 3.91VDD95 pKa = 3.47TTVEE99 pKa = 4.26DD100 pKa = 3.52AKK102 pKa = 10.89RR103 pKa = 11.84WIDD106 pKa = 3.36EE107 pKa = 4.17ATEE110 pKa = 3.77NGYY113 pKa = 10.35DD114 pKa = 3.63VIASYY119 pKa = 10.32HH120 pKa = 6.51HH121 pKa = 6.52YY122 pKa = 9.36PDD124 pKa = 4.38NGSSQASALQHH135 pKa = 6.79AADD138 pKa = 4.69FWVEE142 pKa = 3.86HH143 pKa = 6.43YY144 pKa = 7.72GTLSQDD150 pKa = 3.25SAFTINLMNEE160 pKa = 3.94WGDD163 pKa = 3.91HH164 pKa = 5.12NVTASAYY171 pKa = 9.79ASAYY175 pKa = 10.11NDD177 pKa = 4.36AIDD180 pKa = 3.89TVRR183 pKa = 11.84SGTSYY188 pKa = 11.06DD189 pKa = 3.64GEE191 pKa = 4.49IVCDD195 pKa = 3.73APGWGQGTHH204 pKa = 6.47RR205 pKa = 11.84LADD208 pKa = 3.59AVEE211 pKa = 4.84EE212 pKa = 4.36IDD214 pKa = 6.11DD215 pKa = 4.71DD216 pKa = 4.75DD217 pKa = 5.1LVLSVHH223 pKa = 6.51VYY225 pKa = 9.71PNAYY229 pKa = 9.96NATTGEE235 pKa = 4.16WLRR238 pKa = 11.84PEE240 pKa = 4.29HH241 pKa = 7.01LDD243 pKa = 4.07VIDD246 pKa = 3.72EE247 pKa = 4.26TGYY250 pKa = 9.86PCMIGEE256 pKa = 4.55FGKK259 pKa = 10.59YY260 pKa = 7.64WPTQYY265 pKa = 11.36SDD267 pKa = 3.18TADD270 pKa = 4.2ADD272 pKa = 3.39WLAIVDD278 pKa = 4.09HH279 pKa = 6.32ATSLGWPVIGWAWNGDD295 pKa = 3.63TNEE298 pKa = 3.72QRR300 pKa = 11.84MNMVEE305 pKa = 4.5PYY307 pKa = 9.87WDD309 pKa = 4.22DD310 pKa = 3.57EE311 pKa = 4.64CGGPYY316 pKa = 9.71SEE318 pKa = 5.15SSYY321 pKa = 11.22FDD323 pKa = 3.41VIYY326 pKa = 10.53EE327 pKa = 4.09KK328 pKa = 10.84LGSGGNDD335 pKa = 2.93TGRR338 pKa = 11.84NDD340 pKa = 5.25DD341 pKa = 5.48DD342 pKa = 5.04GDD344 pKa = 4.07EE345 pKa = 4.42NGGNSEE351 pKa = 3.83PTAAIDD357 pKa = 4.5ADD359 pKa = 3.81ATDD362 pKa = 3.86VSVGDD367 pKa = 4.05TPSFDD372 pKa = 3.72ASASSEE378 pKa = 3.81EE379 pKa = 4.35DD380 pKa = 3.22GTIEE384 pKa = 4.17RR385 pKa = 11.84YY386 pKa = 8.96EE387 pKa = 3.91WTFGDD392 pKa = 4.09GTSATGKK399 pKa = 10.09RR400 pKa = 11.84VDD402 pKa = 3.77HH403 pKa = 6.53AFEE406 pKa = 4.4EE407 pKa = 4.76PGEE410 pKa = 4.16YY411 pKa = 9.23TVLLTVTDD419 pKa = 4.77DD420 pKa = 3.91EE421 pKa = 4.85NATDD425 pKa = 3.71TDD427 pKa = 4.54TITVTVTDD435 pKa = 4.08SAKK438 pKa = 10.29SAPGAPANLTVVATTTSSLTIEE460 pKa = 4.05WDD462 pKa = 3.69GVDD465 pKa = 3.48DD466 pKa = 4.32ADD468 pKa = 4.24YY469 pKa = 11.3YY470 pKa = 10.78VVSVDD475 pKa = 3.44GSVDD479 pKa = 3.2HH480 pKa = 6.4EE481 pKa = 4.66TPEE484 pKa = 4.04TTATIDD490 pKa = 3.47EE491 pKa = 5.09LEE493 pKa = 4.33TEE495 pKa = 4.14TGYY498 pKa = 11.04EE499 pKa = 3.92IGVSAVDD506 pKa = 3.27NGGAEE511 pKa = 4.4SATKK515 pKa = 9.43TVSATTASDD524 pKa = 3.67DD525 pKa = 4.64GSDD528 pKa = 4.26EE529 pKa = 4.63DD530 pKa = 6.22DD531 pKa = 3.47EE532 pKa = 5.74DD533 pKa = 4.59RR534 pKa = 11.84GNGDD538 pKa = 3.2FDD540 pKa = 5.37GDD542 pKa = 3.71LVAEE546 pKa = 4.68IRR548 pKa = 11.84ASTTSAWVGEE558 pKa = 4.48HH559 pKa = 6.18IGFFAVDD566 pKa = 3.5RR567 pKa = 11.84TGDD570 pKa = 3.86DD571 pKa = 3.13TWPTEE576 pKa = 4.07LDD578 pKa = 2.99WDD580 pKa = 4.47LGDD583 pKa = 3.97GTTASGWYY591 pKa = 7.37TAYY594 pKa = 10.39AYY596 pKa = 8.85DD597 pKa = 3.76SPGTYY602 pKa = 8.69TVALTATDD610 pKa = 4.41DD611 pKa = 3.62EE612 pKa = 5.22GTTTTHH618 pKa = 5.51EE619 pKa = 3.94VEE621 pKa = 3.83IAVFRR626 pKa = 11.84VRR628 pKa = 11.84RR629 pKa = 11.84SSPAVRR635 pKa = 11.84SSS637 pKa = 2.74
MM1 pKa = 6.59VTTGAWGGFHH11 pKa = 7.67RR12 pKa = 11.84MGCITRR18 pKa = 11.84RR19 pKa = 11.84DD20 pKa = 3.48ILKK23 pKa = 10.38ASLGFGTVMGVGLVGRR39 pKa = 11.84WPEE42 pKa = 4.1TTTSEE47 pKa = 4.19TSFGDD52 pKa = 3.83GVNLQPSYY60 pKa = 10.89FCSANQDD67 pKa = 3.39IGWDD71 pKa = 3.85FMSQYY76 pKa = 11.42SDD78 pKa = 2.75IQTLRR83 pKa = 11.84MRR85 pKa = 11.84IEE87 pKa = 4.06PFSFSEE93 pKa = 3.91VDD95 pKa = 3.47TTVEE99 pKa = 4.26DD100 pKa = 3.52AKK102 pKa = 10.89RR103 pKa = 11.84WIDD106 pKa = 3.36EE107 pKa = 4.17ATEE110 pKa = 3.77NGYY113 pKa = 10.35DD114 pKa = 3.63VIASYY119 pKa = 10.32HH120 pKa = 6.51HH121 pKa = 6.52YY122 pKa = 9.36PDD124 pKa = 4.38NGSSQASALQHH135 pKa = 6.79AADD138 pKa = 4.69FWVEE142 pKa = 3.86HH143 pKa = 6.43YY144 pKa = 7.72GTLSQDD150 pKa = 3.25SAFTINLMNEE160 pKa = 3.94WGDD163 pKa = 3.91HH164 pKa = 5.12NVTASAYY171 pKa = 9.79ASAYY175 pKa = 10.11NDD177 pKa = 4.36AIDD180 pKa = 3.89TVRR183 pKa = 11.84SGTSYY188 pKa = 11.06DD189 pKa = 3.64GEE191 pKa = 4.49IVCDD195 pKa = 3.73APGWGQGTHH204 pKa = 6.47RR205 pKa = 11.84LADD208 pKa = 3.59AVEE211 pKa = 4.84EE212 pKa = 4.36IDD214 pKa = 6.11DD215 pKa = 4.71DD216 pKa = 4.75DD217 pKa = 5.1LVLSVHH223 pKa = 6.51VYY225 pKa = 9.71PNAYY229 pKa = 9.96NATTGEE235 pKa = 4.16WLRR238 pKa = 11.84PEE240 pKa = 4.29HH241 pKa = 7.01LDD243 pKa = 4.07VIDD246 pKa = 3.72EE247 pKa = 4.26TGYY250 pKa = 9.86PCMIGEE256 pKa = 4.55FGKK259 pKa = 10.59YY260 pKa = 7.64WPTQYY265 pKa = 11.36SDD267 pKa = 3.18TADD270 pKa = 4.2ADD272 pKa = 3.39WLAIVDD278 pKa = 4.09HH279 pKa = 6.32ATSLGWPVIGWAWNGDD295 pKa = 3.63TNEE298 pKa = 3.72QRR300 pKa = 11.84MNMVEE305 pKa = 4.5PYY307 pKa = 9.87WDD309 pKa = 4.22DD310 pKa = 3.57EE311 pKa = 4.64CGGPYY316 pKa = 9.71SEE318 pKa = 5.15SSYY321 pKa = 11.22FDD323 pKa = 3.41VIYY326 pKa = 10.53EE327 pKa = 4.09KK328 pKa = 10.84LGSGGNDD335 pKa = 2.93TGRR338 pKa = 11.84NDD340 pKa = 5.25DD341 pKa = 5.48DD342 pKa = 5.04GDD344 pKa = 4.07EE345 pKa = 4.42NGGNSEE351 pKa = 3.83PTAAIDD357 pKa = 4.5ADD359 pKa = 3.81ATDD362 pKa = 3.86VSVGDD367 pKa = 4.05TPSFDD372 pKa = 3.72ASASSEE378 pKa = 3.81EE379 pKa = 4.35DD380 pKa = 3.22GTIEE384 pKa = 4.17RR385 pKa = 11.84YY386 pKa = 8.96EE387 pKa = 3.91WTFGDD392 pKa = 4.09GTSATGKK399 pKa = 10.09RR400 pKa = 11.84VDD402 pKa = 3.77HH403 pKa = 6.53AFEE406 pKa = 4.4EE407 pKa = 4.76PGEE410 pKa = 4.16YY411 pKa = 9.23TVLLTVTDD419 pKa = 4.77DD420 pKa = 3.91EE421 pKa = 4.85NATDD425 pKa = 3.71TDD427 pKa = 4.54TITVTVTDD435 pKa = 4.08SAKK438 pKa = 10.29SAPGAPANLTVVATTTSSLTIEE460 pKa = 4.05WDD462 pKa = 3.69GVDD465 pKa = 3.48DD466 pKa = 4.32ADD468 pKa = 4.24YY469 pKa = 11.3YY470 pKa = 10.78VVSVDD475 pKa = 3.44GSVDD479 pKa = 3.2HH480 pKa = 6.4EE481 pKa = 4.66TPEE484 pKa = 4.04TTATIDD490 pKa = 3.47EE491 pKa = 5.09LEE493 pKa = 4.33TEE495 pKa = 4.14TGYY498 pKa = 11.04EE499 pKa = 3.92IGVSAVDD506 pKa = 3.27NGGAEE511 pKa = 4.4SATKK515 pKa = 9.43TVSATTASDD524 pKa = 3.67DD525 pKa = 4.64GSDD528 pKa = 4.26EE529 pKa = 4.63DD530 pKa = 6.22DD531 pKa = 3.47EE532 pKa = 5.74DD533 pKa = 4.59RR534 pKa = 11.84GNGDD538 pKa = 3.2FDD540 pKa = 5.37GDD542 pKa = 3.71LVAEE546 pKa = 4.68IRR548 pKa = 11.84ASTTSAWVGEE558 pKa = 4.48HH559 pKa = 6.18IGFFAVDD566 pKa = 3.5RR567 pKa = 11.84TGDD570 pKa = 3.86DD571 pKa = 3.13TWPTEE576 pKa = 4.07LDD578 pKa = 2.99WDD580 pKa = 4.47LGDD583 pKa = 3.97GTTASGWYY591 pKa = 7.37TAYY594 pKa = 10.39AYY596 pKa = 8.85DD597 pKa = 3.76SPGTYY602 pKa = 8.69TVALTATDD610 pKa = 4.41DD611 pKa = 3.62EE612 pKa = 5.22GTTTTHH618 pKa = 5.51EE619 pKa = 3.94VEE621 pKa = 3.83IAVFRR626 pKa = 11.84VRR628 pKa = 11.84RR629 pKa = 11.84SSPAVRR635 pKa = 11.84SSS637 pKa = 2.74
Molecular weight: 68.91 kDa
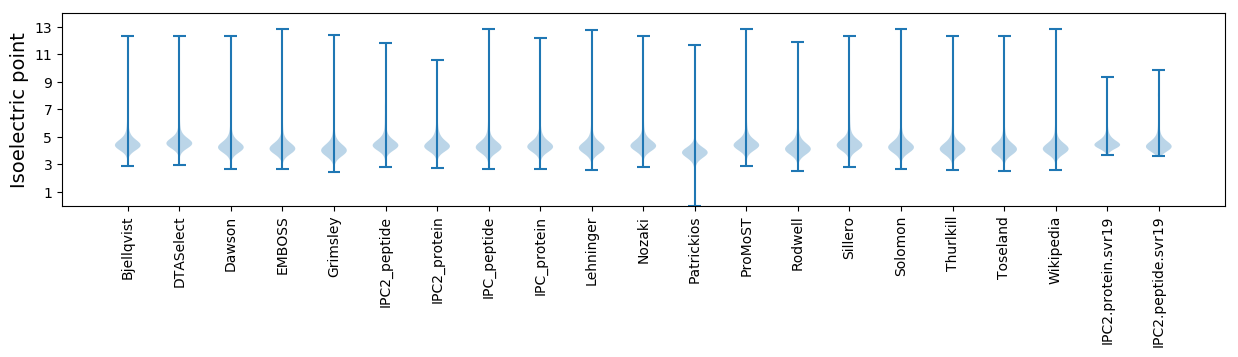
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7D5GL18|A0A7D5GL18_9EURY Arginine decarboxylase OS=Haloterrigena sp. YPL8 OX=1699371 GN=HYG82_11880 PE=3 SV=1
MM1 pKa = 7.57ARR3 pKa = 11.84RR4 pKa = 11.84AVARR8 pKa = 11.84YY9 pKa = 9.38SSTFSKK15 pKa = 10.36RR16 pKa = 11.84RR17 pKa = 11.84YY18 pKa = 7.54TLHH21 pKa = 4.79QHH23 pKa = 7.27IILLCLTVRR32 pKa = 11.84KK33 pKa = 8.33NTTYY37 pKa = 11.35RR38 pKa = 11.84MLLDD42 pKa = 4.22EE43 pKa = 5.71LIEE46 pKa = 3.96MPRR49 pKa = 11.84IRR51 pKa = 11.84RR52 pKa = 11.84AIEE55 pKa = 4.03LEE57 pKa = 4.05EE58 pKa = 4.56LPSPSTLCKK67 pKa = 10.35AFNRR71 pKa = 11.84LGMAVWRR78 pKa = 11.84ILLTLSVTLLPTNGVVGIDD97 pKa = 3.05ASGFDD102 pKa = 3.53RR103 pKa = 11.84SHH105 pKa = 6.94ASKK108 pKa = 10.5HH109 pKa = 3.65YY110 pKa = 8.79TKK112 pKa = 9.81RR113 pKa = 11.84TKK115 pKa = 9.96LTIQQLTVTLLVDD128 pKa = 3.67TRR130 pKa = 11.84ANAILDD136 pKa = 3.56LHH138 pKa = 5.57VTTTRR143 pKa = 11.84THH145 pKa = 6.9DD146 pKa = 3.68SKK148 pKa = 10.95IAPSLSKK155 pKa = 10.73RR156 pKa = 11.84NSGEE160 pKa = 3.76VAIFLGDD167 pKa = 4.35KK168 pKa = 10.91GYY170 pKa = 10.96DD171 pKa = 3.48DD172 pKa = 4.37QKK174 pKa = 10.85IHH176 pKa = 6.93ALAHH180 pKa = 6.05EE181 pKa = 5.01DD182 pKa = 3.73GVRR185 pKa = 11.84PLIKK189 pKa = 10.1HH190 pKa = 6.19RR191 pKa = 11.84EE192 pKa = 3.73FSSLHH197 pKa = 5.64KK198 pKa = 10.65ARR200 pKa = 11.84NARR203 pKa = 11.84LDD205 pKa = 3.64ADD207 pKa = 3.98SYY209 pKa = 9.04GHH211 pKa = 5.2VVRR214 pKa = 11.84TRR216 pKa = 11.84RR217 pKa = 11.84RR218 pKa = 11.84TLASNGNTAHH228 pKa = 7.0SSAHH232 pKa = 5.68DD233 pKa = 3.57TGGSSSVNSLSAVSLTPSTRR253 pKa = 11.84HH254 pKa = 5.97PEE256 pKa = 3.74RR257 pKa = 5.77
MM1 pKa = 7.57ARR3 pKa = 11.84RR4 pKa = 11.84AVARR8 pKa = 11.84YY9 pKa = 9.38SSTFSKK15 pKa = 10.36RR16 pKa = 11.84RR17 pKa = 11.84YY18 pKa = 7.54TLHH21 pKa = 4.79QHH23 pKa = 7.27IILLCLTVRR32 pKa = 11.84KK33 pKa = 8.33NTTYY37 pKa = 11.35RR38 pKa = 11.84MLLDD42 pKa = 4.22EE43 pKa = 5.71LIEE46 pKa = 3.96MPRR49 pKa = 11.84IRR51 pKa = 11.84RR52 pKa = 11.84AIEE55 pKa = 4.03LEE57 pKa = 4.05EE58 pKa = 4.56LPSPSTLCKK67 pKa = 10.35AFNRR71 pKa = 11.84LGMAVWRR78 pKa = 11.84ILLTLSVTLLPTNGVVGIDD97 pKa = 3.05ASGFDD102 pKa = 3.53RR103 pKa = 11.84SHH105 pKa = 6.94ASKK108 pKa = 10.5HH109 pKa = 3.65YY110 pKa = 8.79TKK112 pKa = 9.81RR113 pKa = 11.84TKK115 pKa = 9.96LTIQQLTVTLLVDD128 pKa = 3.67TRR130 pKa = 11.84ANAILDD136 pKa = 3.56LHH138 pKa = 5.57VTTTRR143 pKa = 11.84THH145 pKa = 6.9DD146 pKa = 3.68SKK148 pKa = 10.95IAPSLSKK155 pKa = 10.73RR156 pKa = 11.84NSGEE160 pKa = 3.76VAIFLGDD167 pKa = 4.35KK168 pKa = 10.91GYY170 pKa = 10.96DD171 pKa = 3.48DD172 pKa = 4.37QKK174 pKa = 10.85IHH176 pKa = 6.93ALAHH180 pKa = 6.05EE181 pKa = 5.01DD182 pKa = 3.73GVRR185 pKa = 11.84PLIKK189 pKa = 10.1HH190 pKa = 6.19RR191 pKa = 11.84EE192 pKa = 3.73FSSLHH197 pKa = 5.64KK198 pKa = 10.65ARR200 pKa = 11.84NARR203 pKa = 11.84LDD205 pKa = 3.64ADD207 pKa = 3.98SYY209 pKa = 9.04GHH211 pKa = 5.2VVRR214 pKa = 11.84TRR216 pKa = 11.84RR217 pKa = 11.84RR218 pKa = 11.84TLASNGNTAHH228 pKa = 7.0SSAHH232 pKa = 5.68DD233 pKa = 3.57TGGSSSVNSLSAVSLTPSTRR253 pKa = 11.84HH254 pKa = 5.97PEE256 pKa = 3.74RR257 pKa = 5.77
Molecular weight: 28.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1144613 |
42 |
1563 |
287.5 |
31.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.052 ± 0.055 | 0.766 ± 0.012 |
8.5 ± 0.052 | 8.601 ± 0.065 |
3.335 ± 0.028 | 8.351 ± 0.045 |
2.007 ± 0.02 | 4.724 ± 0.028 |
1.905 ± 0.021 | 8.698 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.754 ± 0.015 | 2.601 ± 0.022 |
4.556 ± 0.028 | 2.56 ± 0.024 |
6.511 ± 0.035 | 5.947 ± 0.035 |
6.7 ± 0.03 | 8.504 ± 0.041 |
1.18 ± 0.016 | 2.751 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
