
Canis familiaris papillomavirus 13
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Taupapillomavirus; Taupapillomavirus 2
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
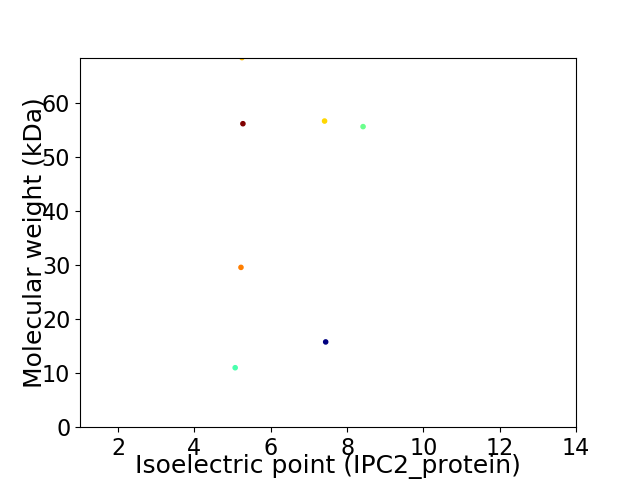
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J7JLS2|J7JLS2_9PAPI Protein E6 OS=Canis familiaris papillomavirus 13 OX=1226723 GN=E6 PE=3 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84GLAPTNRR9 pKa = 11.84DD10 pKa = 2.86LDD12 pKa = 4.11LEE14 pKa = 4.53LGEE17 pKa = 5.31LVLPEE22 pKa = 4.37NLLSSEE28 pKa = 4.11RR29 pKa = 11.84LDD31 pKa = 3.73TEE33 pKa = 4.4EE34 pKa = 5.1EE35 pKa = 4.05EE36 pKa = 4.98SEE38 pKa = 4.7PDD40 pKa = 3.42PQHH43 pKa = 5.82YY44 pKa = 9.53RR45 pKa = 11.84VVTNCGRR52 pKa = 11.84CHH54 pKa = 4.84STLRR58 pKa = 11.84VFVAVLSVFQLRR70 pKa = 11.84TFEE73 pKa = 4.28QLLLDD78 pKa = 4.6GFTIVCLRR86 pKa = 11.84CSQQQSHH93 pKa = 5.88GRR95 pKa = 11.84HH96 pKa = 5.26
MM1 pKa = 7.86RR2 pKa = 11.84GLAPTNRR9 pKa = 11.84DD10 pKa = 2.86LDD12 pKa = 4.11LEE14 pKa = 4.53LGEE17 pKa = 5.31LVLPEE22 pKa = 4.37NLLSSEE28 pKa = 4.11RR29 pKa = 11.84LDD31 pKa = 3.73TEE33 pKa = 4.4EE34 pKa = 5.1EE35 pKa = 4.05EE36 pKa = 4.98SEE38 pKa = 4.7PDD40 pKa = 3.42PQHH43 pKa = 5.82YY44 pKa = 9.53RR45 pKa = 11.84VVTNCGRR52 pKa = 11.84CHH54 pKa = 4.84STLRR58 pKa = 11.84VFVAVLSVFQLRR70 pKa = 11.84TFEE73 pKa = 4.28QLLLDD78 pKa = 4.6GFTIVCLRR86 pKa = 11.84CSQQQSHH93 pKa = 5.88GRR95 pKa = 11.84HH96 pKa = 5.26
Molecular weight: 10.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J7JMM7|J7JMM7_9PAPI Minor capsid protein L2 OS=Canis familiaris papillomavirus 13 OX=1226723 GN=L2 PE=3 SV=1
MM1 pKa = 7.65EE2 pKa = 5.0NLHH5 pKa = 5.89TRR7 pKa = 11.84FDD9 pKa = 3.94VVQEE13 pKa = 3.98VLFQHH18 pKa = 6.29YY19 pKa = 8.74EE20 pKa = 3.94QGSHH24 pKa = 6.76KK25 pKa = 10.68LSDD28 pKa = 3.39HH29 pKa = 6.09ALFWEE34 pKa = 4.79AKK36 pKa = 8.4RR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.15AIMLFFARR47 pKa = 11.84KK48 pKa = 8.68RR49 pKa = 11.84QMPRR53 pKa = 11.84LGFQPVPALSVSEE66 pKa = 5.0GNAKK70 pKa = 10.24DD71 pKa = 3.94AIKK74 pKa = 10.15MGLLLSSLQKK84 pKa = 10.5SPYY87 pKa = 9.85GDD89 pKa = 3.79EE90 pKa = 3.93PWRR93 pKa = 11.84ITDD96 pKa = 3.65VSLEE100 pKa = 3.99MLNTEE105 pKa = 5.16PKK107 pKa = 10.71DD108 pKa = 3.71CFKK111 pKa = 11.37KK112 pKa = 10.58NGITVEE118 pKa = 3.8VHH120 pKa = 6.25YY121 pKa = 11.39DD122 pKa = 3.51NDD124 pKa = 3.99PDD126 pKa = 3.76NAAHH130 pKa = 5.04YY131 pKa = 7.14TSWSEE136 pKa = 3.8IYY138 pKa = 9.52YY139 pKa = 10.66QNVDD143 pKa = 3.78DD144 pKa = 5.17DD145 pKa = 4.15WVKK148 pKa = 10.22VRR150 pKa = 11.84GRR152 pKa = 11.84VNYY155 pKa = 9.74EE156 pKa = 3.32GLFFVDD162 pKa = 3.28EE163 pKa = 4.98DD164 pKa = 4.16GEE166 pKa = 3.95EE167 pKa = 4.0RR168 pKa = 11.84YY169 pKa = 9.65YY170 pKa = 11.61VRR172 pKa = 11.84FQKK175 pKa = 10.64DD176 pKa = 2.98AEE178 pKa = 4.43RR179 pKa = 11.84YY180 pKa = 9.04GVTGMWRR187 pKa = 11.84VHH189 pKa = 5.61YY190 pKa = 9.14KK191 pKa = 10.28QSIISASVSSSGAAPAPSTHH211 pKa = 7.06HH212 pKa = 6.47AQTWWPDD219 pKa = 3.15WPDD222 pKa = 3.38AAPDD226 pKa = 3.62SSTGSTGQRR235 pKa = 11.84SSSSEE240 pKa = 3.76SRR242 pKa = 11.84EE243 pKa = 3.74GSEE246 pKa = 4.92DD247 pKa = 2.75PWEE250 pKa = 4.04RR251 pKa = 11.84RR252 pKa = 11.84EE253 pKa = 3.83RR254 pKa = 11.84RR255 pKa = 11.84LAGTRR260 pKa = 11.84RR261 pKa = 11.84GRR263 pKa = 11.84GGGRR267 pKa = 11.84GGGRR271 pKa = 11.84GRR273 pKa = 11.84GPEE276 pKa = 4.15ASPSPAPTPSALPVQGDD293 pKa = 4.25LPQSRR298 pKa = 11.84QRR300 pKa = 11.84EE301 pKa = 4.19EE302 pKa = 4.07EE303 pKa = 4.14EE304 pKa = 4.41EE305 pKa = 4.08EE306 pKa = 4.56GGPRR310 pKa = 11.84PHH312 pKa = 5.97QACWEE317 pKa = 4.14NGGEE321 pKa = 4.41GPSRR325 pKa = 11.84GSGGIKK331 pKa = 9.97RR332 pKa = 11.84RR333 pKa = 11.84GTGSRR338 pKa = 11.84GRR340 pKa = 11.84PPKK343 pKa = 10.5KK344 pKa = 9.59KK345 pKa = 8.75FCRR348 pKa = 11.84GGRR351 pKa = 11.84QQGEE355 pKa = 4.33FHH357 pKa = 6.5TPSPPTLQSSSFTSPGGDD375 pKa = 3.23FRR377 pKa = 11.84GGPHH381 pKa = 7.41PPRR384 pKa = 11.84LSLTLLDD391 pKa = 4.11GKK393 pKa = 11.12VGGRR397 pKa = 11.84SGQFGGHH404 pKa = 6.92PPQGPGQAPAQAGGFPVIVLKK425 pKa = 11.25GPGNSLKK432 pKa = 10.45CFRR435 pKa = 11.84LRR437 pKa = 11.84CRR439 pKa = 11.84RR440 pKa = 11.84GHH442 pKa = 4.98SQKK445 pKa = 10.23FLCISTGYY453 pKa = 8.19TWVCGEE459 pKa = 3.8GDD461 pKa = 3.18KK462 pKa = 11.15VGRR465 pKa = 11.84QRR467 pKa = 11.84LMIGFRR473 pKa = 11.84DD474 pKa = 3.23EE475 pKa = 4.24GQRR478 pKa = 11.84SFFLKK483 pKa = 10.2NVKK486 pKa = 9.79IPSTFDD492 pKa = 3.09LSFGMFDD499 pKa = 3.64SLL501 pKa = 5.05
MM1 pKa = 7.65EE2 pKa = 5.0NLHH5 pKa = 5.89TRR7 pKa = 11.84FDD9 pKa = 3.94VVQEE13 pKa = 3.98VLFQHH18 pKa = 6.29YY19 pKa = 8.74EE20 pKa = 3.94QGSHH24 pKa = 6.76KK25 pKa = 10.68LSDD28 pKa = 3.39HH29 pKa = 6.09ALFWEE34 pKa = 4.79AKK36 pKa = 8.4RR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.15AIMLFFARR47 pKa = 11.84KK48 pKa = 8.68RR49 pKa = 11.84QMPRR53 pKa = 11.84LGFQPVPALSVSEE66 pKa = 5.0GNAKK70 pKa = 10.24DD71 pKa = 3.94AIKK74 pKa = 10.15MGLLLSSLQKK84 pKa = 10.5SPYY87 pKa = 9.85GDD89 pKa = 3.79EE90 pKa = 3.93PWRR93 pKa = 11.84ITDD96 pKa = 3.65VSLEE100 pKa = 3.99MLNTEE105 pKa = 5.16PKK107 pKa = 10.71DD108 pKa = 3.71CFKK111 pKa = 11.37KK112 pKa = 10.58NGITVEE118 pKa = 3.8VHH120 pKa = 6.25YY121 pKa = 11.39DD122 pKa = 3.51NDD124 pKa = 3.99PDD126 pKa = 3.76NAAHH130 pKa = 5.04YY131 pKa = 7.14TSWSEE136 pKa = 3.8IYY138 pKa = 9.52YY139 pKa = 10.66QNVDD143 pKa = 3.78DD144 pKa = 5.17DD145 pKa = 4.15WVKK148 pKa = 10.22VRR150 pKa = 11.84GRR152 pKa = 11.84VNYY155 pKa = 9.74EE156 pKa = 3.32GLFFVDD162 pKa = 3.28EE163 pKa = 4.98DD164 pKa = 4.16GEE166 pKa = 3.95EE167 pKa = 4.0RR168 pKa = 11.84YY169 pKa = 9.65YY170 pKa = 11.61VRR172 pKa = 11.84FQKK175 pKa = 10.64DD176 pKa = 2.98AEE178 pKa = 4.43RR179 pKa = 11.84YY180 pKa = 9.04GVTGMWRR187 pKa = 11.84VHH189 pKa = 5.61YY190 pKa = 9.14KK191 pKa = 10.28QSIISASVSSSGAAPAPSTHH211 pKa = 7.06HH212 pKa = 6.47AQTWWPDD219 pKa = 3.15WPDD222 pKa = 3.38AAPDD226 pKa = 3.62SSTGSTGQRR235 pKa = 11.84SSSSEE240 pKa = 3.76SRR242 pKa = 11.84EE243 pKa = 3.74GSEE246 pKa = 4.92DD247 pKa = 2.75PWEE250 pKa = 4.04RR251 pKa = 11.84RR252 pKa = 11.84EE253 pKa = 3.83RR254 pKa = 11.84RR255 pKa = 11.84LAGTRR260 pKa = 11.84RR261 pKa = 11.84GRR263 pKa = 11.84GGGRR267 pKa = 11.84GGGRR271 pKa = 11.84GRR273 pKa = 11.84GPEE276 pKa = 4.15ASPSPAPTPSALPVQGDD293 pKa = 4.25LPQSRR298 pKa = 11.84QRR300 pKa = 11.84EE301 pKa = 4.19EE302 pKa = 4.07EE303 pKa = 4.14EE304 pKa = 4.41EE305 pKa = 4.08EE306 pKa = 4.56GGPRR310 pKa = 11.84PHH312 pKa = 5.97QACWEE317 pKa = 4.14NGGEE321 pKa = 4.41GPSRR325 pKa = 11.84GSGGIKK331 pKa = 9.97RR332 pKa = 11.84RR333 pKa = 11.84GTGSRR338 pKa = 11.84GRR340 pKa = 11.84PPKK343 pKa = 10.5KK344 pKa = 9.59KK345 pKa = 8.75FCRR348 pKa = 11.84GGRR351 pKa = 11.84QQGEE355 pKa = 4.33FHH357 pKa = 6.5TPSPPTLQSSSFTSPGGDD375 pKa = 3.23FRR377 pKa = 11.84GGPHH381 pKa = 7.41PPRR384 pKa = 11.84LSLTLLDD391 pKa = 4.11GKK393 pKa = 11.12VGGRR397 pKa = 11.84SGQFGGHH404 pKa = 6.92PPQGPGQAPAQAGGFPVIVLKK425 pKa = 11.25GPGNSLKK432 pKa = 10.45CFRR435 pKa = 11.84LRR437 pKa = 11.84CRR439 pKa = 11.84RR440 pKa = 11.84GHH442 pKa = 4.98SQKK445 pKa = 10.23FLCISTGYY453 pKa = 8.19TWVCGEE459 pKa = 3.8GDD461 pKa = 3.18KK462 pKa = 11.15VGRR465 pKa = 11.84QRR467 pKa = 11.84LMIGFRR473 pKa = 11.84DD474 pKa = 3.23EE475 pKa = 4.24GQRR478 pKa = 11.84SFFLKK483 pKa = 10.2NVKK486 pKa = 9.79IPSTFDD492 pKa = 3.09LSFGMFDD499 pKa = 3.64SLL501 pKa = 5.05
Molecular weight: 55.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2639 |
96 |
604 |
377.0 |
41.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.267 ± 0.412 | 2.387 ± 0.709 |
6.631 ± 0.487 | 6.328 ± 0.649 |
4.888 ± 0.257 | 8.867 ± 1.121 |
2.728 ± 0.263 | 2.804 ± 0.287 |
4.888 ± 0.695 | 8.147 ± 0.595 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.743 ± 0.294 | 3.297 ± 0.65 |
7.768 ± 1.3 | 3.789 ± 0.486 |
6.859 ± 0.732 | 8.45 ± 0.66 |
5.532 ± 0.668 | 6.063 ± 0.81 |
1.402 ± 0.305 | 2.16 ± 0.355 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
