
Paenibacillaceae bacterium
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; unclassified Paenibacillaceae
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
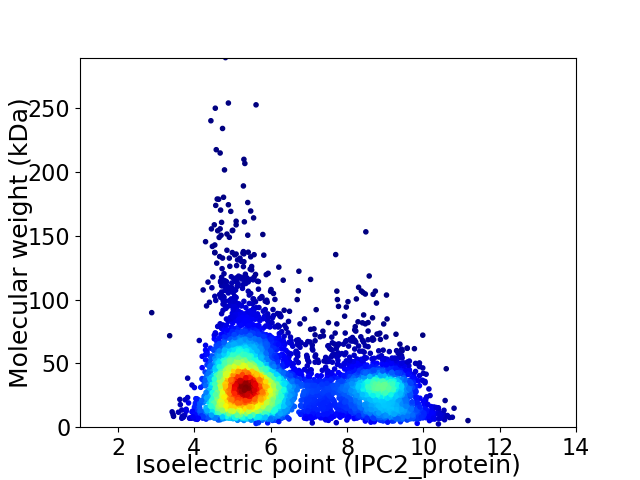
Virtual 2D-PAGE plot for 6717 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q2LXI0|A0A4Q2LXI0_9BACL Transcriptional regulator OS=Paenibacillaceae bacterium OX=2003592 GN=EBB07_03410 PE=4 SV=1
MM1 pKa = 7.8LKK3 pKa = 10.23RR4 pKa = 11.84NKK6 pKa = 9.56IKK8 pKa = 10.67LVAVMLMLGALFLAGCSEE26 pKa = 3.85WLEE29 pKa = 4.31EE30 pKa = 4.91NSEE33 pKa = 4.0LWEE36 pKa = 4.08EE37 pKa = 4.65AEE39 pKa = 4.32LQAAADD45 pKa = 3.76NDD47 pKa = 4.03EE48 pKa = 4.41EE49 pKa = 5.04NGLSSVEE56 pKa = 3.87TDD58 pKa = 3.62CYY60 pKa = 10.92VDD62 pKa = 4.37EE63 pKa = 5.55IYY65 pKa = 10.85LSEE68 pKa = 4.23EE69 pKa = 4.09SRR71 pKa = 11.84CITEE75 pKa = 4.05WTCDD79 pKa = 3.68DD80 pKa = 5.52LEE82 pKa = 4.58SCEE85 pKa = 4.1NLGNEE90 pKa = 4.46LMSNLIEE97 pKa = 4.43NYY99 pKa = 10.11GSFVNLEE106 pKa = 3.77EE107 pKa = 4.98SEE109 pKa = 4.09IEE111 pKa = 4.17EE112 pKa = 4.15NEE114 pKa = 4.03KK115 pKa = 10.93LSALEE120 pKa = 4.04EE121 pKa = 4.15QPKK124 pKa = 9.83ASYY127 pKa = 8.89WVEE130 pKa = 3.86EE131 pKa = 4.29EE132 pKa = 4.26EE133 pKa = 5.73LVLEE137 pKa = 5.55DD138 pKa = 4.43GDD140 pKa = 4.06EE141 pKa = 4.54SDD143 pKa = 4.44DD144 pKa = 5.32HH145 pKa = 7.59RR146 pKa = 11.84EE147 pKa = 3.77LWEE150 pKa = 4.37GFAAIIPPHH159 pKa = 5.81EE160 pKa = 4.84RR161 pKa = 11.84EE162 pKa = 4.38DD163 pKa = 3.54VTLFTVFSHH172 pKa = 6.64YY173 pKa = 9.01DD174 pKa = 3.05TLGFVRR180 pKa = 11.84QNDD183 pKa = 3.84DD184 pKa = 5.55DD185 pKa = 4.98YY186 pKa = 11.42STWTFGLNIEE196 pKa = 4.73GAASLNEE203 pKa = 3.77AVNTNIHH210 pKa = 5.96EE211 pKa = 4.71FGHH214 pKa = 6.96LLTLRR219 pKa = 11.84TQQVDD224 pKa = 3.43PDD226 pKa = 4.19AEE228 pKa = 4.01EE229 pKa = 4.52HH230 pKa = 6.48LCNTYY235 pKa = 10.8YY236 pKa = 10.99VDD238 pKa = 4.78DD239 pKa = 4.71GCAEE243 pKa = 3.95SDD245 pKa = 3.29AYY247 pKa = 9.87IYY249 pKa = 11.18AFYY252 pKa = 10.13QQFWSDD258 pKa = 3.63DD259 pKa = 3.71QQDD262 pKa = 3.46EE263 pKa = 4.73DD264 pKa = 3.93EE265 pKa = 5.19DD266 pKa = 4.89SFVSEE271 pKa = 4.22YY272 pKa = 11.37ALTNILEE279 pKa = 5.15DD280 pKa = 3.87IAEE283 pKa = 4.04SWAFFVLADD292 pKa = 4.57RR293 pKa = 11.84PDD295 pKa = 3.69GDD297 pKa = 3.8TVVEE301 pKa = 3.87QKK303 pKa = 10.55ILFFYY308 pKa = 10.3EE309 pKa = 3.86YY310 pKa = 10.69EE311 pKa = 4.1EE312 pKa = 4.16LVKK315 pKa = 10.9LRR317 pKa = 11.84TEE319 pKa = 3.97LLARR323 pKa = 11.84ITSWRR328 pKa = 11.84EE329 pKa = 3.17RR330 pKa = 11.84AASS333 pKa = 3.33
MM1 pKa = 7.8LKK3 pKa = 10.23RR4 pKa = 11.84NKK6 pKa = 9.56IKK8 pKa = 10.67LVAVMLMLGALFLAGCSEE26 pKa = 3.85WLEE29 pKa = 4.31EE30 pKa = 4.91NSEE33 pKa = 4.0LWEE36 pKa = 4.08EE37 pKa = 4.65AEE39 pKa = 4.32LQAAADD45 pKa = 3.76NDD47 pKa = 4.03EE48 pKa = 4.41EE49 pKa = 5.04NGLSSVEE56 pKa = 3.87TDD58 pKa = 3.62CYY60 pKa = 10.92VDD62 pKa = 4.37EE63 pKa = 5.55IYY65 pKa = 10.85LSEE68 pKa = 4.23EE69 pKa = 4.09SRR71 pKa = 11.84CITEE75 pKa = 4.05WTCDD79 pKa = 3.68DD80 pKa = 5.52LEE82 pKa = 4.58SCEE85 pKa = 4.1NLGNEE90 pKa = 4.46LMSNLIEE97 pKa = 4.43NYY99 pKa = 10.11GSFVNLEE106 pKa = 3.77EE107 pKa = 4.98SEE109 pKa = 4.09IEE111 pKa = 4.17EE112 pKa = 4.15NEE114 pKa = 4.03KK115 pKa = 10.93LSALEE120 pKa = 4.04EE121 pKa = 4.15QPKK124 pKa = 9.83ASYY127 pKa = 8.89WVEE130 pKa = 3.86EE131 pKa = 4.29EE132 pKa = 4.26EE133 pKa = 5.73LVLEE137 pKa = 5.55DD138 pKa = 4.43GDD140 pKa = 4.06EE141 pKa = 4.54SDD143 pKa = 4.44DD144 pKa = 5.32HH145 pKa = 7.59RR146 pKa = 11.84EE147 pKa = 3.77LWEE150 pKa = 4.37GFAAIIPPHH159 pKa = 5.81EE160 pKa = 4.84RR161 pKa = 11.84EE162 pKa = 4.38DD163 pKa = 3.54VTLFTVFSHH172 pKa = 6.64YY173 pKa = 9.01DD174 pKa = 3.05TLGFVRR180 pKa = 11.84QNDD183 pKa = 3.84DD184 pKa = 5.55DD185 pKa = 4.98YY186 pKa = 11.42STWTFGLNIEE196 pKa = 4.73GAASLNEE203 pKa = 3.77AVNTNIHH210 pKa = 5.96EE211 pKa = 4.71FGHH214 pKa = 6.96LLTLRR219 pKa = 11.84TQQVDD224 pKa = 3.43PDD226 pKa = 4.19AEE228 pKa = 4.01EE229 pKa = 4.52HH230 pKa = 6.48LCNTYY235 pKa = 10.8YY236 pKa = 10.99VDD238 pKa = 4.78DD239 pKa = 4.71GCAEE243 pKa = 3.95SDD245 pKa = 3.29AYY247 pKa = 9.87IYY249 pKa = 11.18AFYY252 pKa = 10.13QQFWSDD258 pKa = 3.63DD259 pKa = 3.71QQDD262 pKa = 3.46EE263 pKa = 4.73DD264 pKa = 3.93EE265 pKa = 5.19DD266 pKa = 4.89SFVSEE271 pKa = 4.22YY272 pKa = 11.37ALTNILEE279 pKa = 5.15DD280 pKa = 3.87IAEE283 pKa = 4.04SWAFFVLADD292 pKa = 4.57RR293 pKa = 11.84PDD295 pKa = 3.69GDD297 pKa = 3.8TVVEE301 pKa = 3.87QKK303 pKa = 10.55ILFFYY308 pKa = 10.3EE309 pKa = 3.86YY310 pKa = 10.69EE311 pKa = 4.1EE312 pKa = 4.16LVKK315 pKa = 10.9LRR317 pKa = 11.84TEE319 pKa = 3.97LLARR323 pKa = 11.84ITSWRR328 pKa = 11.84EE329 pKa = 3.17RR330 pKa = 11.84AASS333 pKa = 3.33
Molecular weight: 38.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q2M2V6|A0A4Q2M2V6_9BACL Gfo/Idh/MocA family oxidoreductase OS=Paenibacillaceae bacterium OX=2003592 GN=EBB07_03175 PE=4 SV=1
MM1 pKa = 7.82RR2 pKa = 11.84PTFRR6 pKa = 11.84PNVSKK11 pKa = 10.72RR12 pKa = 11.84KK13 pKa = 8.96KK14 pKa = 8.25VHH16 pKa = 5.49GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 9.99NGRR28 pKa = 11.84KK29 pKa = 8.83VLSARR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.26GRR39 pKa = 11.84KK40 pKa = 8.56VLSAA44 pKa = 4.05
MM1 pKa = 7.82RR2 pKa = 11.84PTFRR6 pKa = 11.84PNVSKK11 pKa = 10.72RR12 pKa = 11.84KK13 pKa = 8.96KK14 pKa = 8.25VHH16 pKa = 5.49GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 9.99NGRR28 pKa = 11.84KK29 pKa = 8.83VLSARR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.26GRR39 pKa = 11.84KK40 pKa = 8.56VLSAA44 pKa = 4.05
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2160698 |
19 |
2710 |
321.7 |
35.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.461 ± 0.039 | 0.765 ± 0.01 |
5.171 ± 0.025 | 6.749 ± 0.038 |
4.15 ± 0.023 | 7.24 ± 0.033 |
2.139 ± 0.016 | 6.756 ± 0.03 |
5.134 ± 0.03 | 10.166 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.774 ± 0.015 | 4.0 ± 0.027 |
3.92 ± 0.017 | 4.132 ± 0.021 |
5.104 ± 0.03 | 6.305 ± 0.023 |
5.282 ± 0.026 | 6.842 ± 0.027 |
1.338 ± 0.013 | 3.57 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
