
Roseobacter phage CRP-6
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; unclassified Caudovirales
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
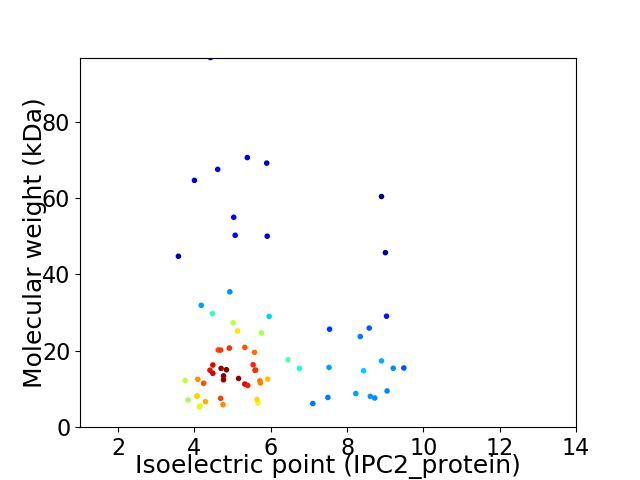
Virtual 2D-PAGE plot for 69 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A646QWK5|A0A646QWK5_9CAUD Putative tail structural protein OS=Roseobacter phage CRP-6 OX=2559285 GN=CRP6_gp46 PE=4 SV=1
MM1 pKa = 7.55ALVLKK6 pKa = 10.69DD7 pKa = 3.59RR8 pKa = 11.84VKK10 pKa = 10.36EE11 pKa = 4.29TTTTTGTGTYY21 pKa = 8.78TLAGAEE27 pKa = 4.68DD28 pKa = 4.41GFQAFSAIGDD38 pKa = 3.96GNITYY43 pKa = 7.8YY44 pKa = 10.45TATNAAGDD52 pKa = 3.91WEE54 pKa = 4.63VGIGTYY60 pKa = 7.76TASGTTLARR69 pKa = 11.84TTILSSSNGGSAVDD83 pKa = 3.31WSAGEE88 pKa = 3.91KK89 pKa = 9.91QVFVTQPASKK99 pKa = 10.5ASYY102 pKa = 8.46VDD104 pKa = 3.44EE105 pKa = 4.81NGFHH109 pKa = 7.12IGNIFEE115 pKa = 4.59KK116 pKa = 10.55CVEE119 pKa = 4.24LKK121 pKa = 8.41TTVANKK127 pKa = 8.99PAYY130 pKa = 9.42EE131 pKa = 4.1EE132 pKa = 4.45GRR134 pKa = 11.84LFYY137 pKa = 11.04DD138 pKa = 3.44KK139 pKa = 11.44AFGALAFYY147 pKa = 10.81NDD149 pKa = 3.92EE150 pKa = 4.42SDD152 pKa = 2.95ITLQIGQEE160 pKa = 4.1EE161 pKa = 5.19YY162 pKa = 10.46IRR164 pKa = 11.84VYY166 pKa = 11.17NDD168 pKa = 2.86TGATLLNGTPVYY180 pKa = 9.92LTGEE184 pKa = 4.22SGATPTVSAARR195 pKa = 11.84ADD197 pKa = 3.56TTYY200 pKa = 11.4AEE202 pKa = 4.29SQAVGVTTHH211 pKa = 7.36DD212 pKa = 3.77IEE214 pKa = 4.83NNTIGYY220 pKa = 6.12VTVRR224 pKa = 11.84GLIADD229 pKa = 3.75VDD231 pKa = 4.26TSHH234 pKa = 6.66LTVGEE239 pKa = 4.17PVHH242 pKa = 5.99VAVGGGTVTEE252 pKa = 4.55SPSYY256 pKa = 9.94PYY258 pKa = 10.78YY259 pKa = 7.51PTEE262 pKa = 3.95VGVCLISAAIGGCIYY277 pKa = 10.91VSISSEE283 pKa = 4.05TFQTMRR289 pKa = 11.84VDD291 pKa = 3.88GNSHH295 pKa = 6.63FDD297 pKa = 3.47ADD299 pKa = 3.87VTIDD303 pKa = 3.41GDD305 pKa = 4.14LVVNGTQTITSSNNIALSGAFSYY328 pKa = 10.82FNSGDD333 pKa = 3.63TIGADD338 pKa = 3.05NTSFTGTGTNDD349 pKa = 3.16GVFTGHH355 pKa = 6.57YY356 pKa = 10.13NGTSSNKK363 pKa = 8.16TYY365 pKa = 10.48YY366 pKa = 10.65VRR368 pKa = 11.84ISDD371 pKa = 4.16DD372 pKa = 3.02QSAPDD377 pKa = 3.52EE378 pKa = 4.79FEE380 pKa = 3.74WSLDD384 pKa = 3.4NFVTIEE390 pKa = 3.91ATGIAITTDD399 pKa = 3.39DD400 pKa = 3.46QALEE404 pKa = 3.96EE405 pKa = 4.23GVNIKK410 pKa = 10.34FNADD414 pKa = 3.24NGHH417 pKa = 5.73VLGDD421 pKa = 3.7VWSGTASPTNVDD433 pKa = 2.98TGIASNRR440 pKa = 11.84NTGTSGVGYY449 pKa = 6.79THH451 pKa = 6.88VGMYY455 pKa = 10.47YY456 pKa = 10.36DD457 pKa = 3.52VSSNYY462 pKa = 7.02WTFFDD467 pKa = 4.74EE468 pKa = 4.6YY469 pKa = 10.87APEE472 pKa = 4.08PTGTIDD478 pKa = 3.15TSHH481 pKa = 7.0ASFSYY486 pKa = 11.27GDD488 pKa = 3.27IKK490 pKa = 11.15VNSVIGDD497 pKa = 3.52VVGNLTGIASSATQLANNRR516 pKa = 11.84NITLSGDD523 pKa = 3.56VTGTAVFNGGADD535 pKa = 3.66ANITATVVNDD545 pKa = 3.29SHH547 pKa = 6.31THH549 pKa = 4.64DD550 pKa = 3.24TRR552 pKa = 11.84YY553 pKa = 10.15VQLAGDD559 pKa = 4.01TMSGTLNVPTVDD571 pKa = 3.99FGDD574 pKa = 3.55WTITEE579 pKa = 4.57SGGSLYY585 pKa = 10.46FAYY588 pKa = 10.4QGTNKK593 pKa = 10.24LKK595 pKa = 10.69LDD597 pKa = 3.63TSGTLSVTNDD607 pKa = 2.9VQTDD611 pKa = 3.47QTII614 pKa = 3.13
MM1 pKa = 7.55ALVLKK6 pKa = 10.69DD7 pKa = 3.59RR8 pKa = 11.84VKK10 pKa = 10.36EE11 pKa = 4.29TTTTTGTGTYY21 pKa = 8.78TLAGAEE27 pKa = 4.68DD28 pKa = 4.41GFQAFSAIGDD38 pKa = 3.96GNITYY43 pKa = 7.8YY44 pKa = 10.45TATNAAGDD52 pKa = 3.91WEE54 pKa = 4.63VGIGTYY60 pKa = 7.76TASGTTLARR69 pKa = 11.84TTILSSSNGGSAVDD83 pKa = 3.31WSAGEE88 pKa = 3.91KK89 pKa = 9.91QVFVTQPASKK99 pKa = 10.5ASYY102 pKa = 8.46VDD104 pKa = 3.44EE105 pKa = 4.81NGFHH109 pKa = 7.12IGNIFEE115 pKa = 4.59KK116 pKa = 10.55CVEE119 pKa = 4.24LKK121 pKa = 8.41TTVANKK127 pKa = 8.99PAYY130 pKa = 9.42EE131 pKa = 4.1EE132 pKa = 4.45GRR134 pKa = 11.84LFYY137 pKa = 11.04DD138 pKa = 3.44KK139 pKa = 11.44AFGALAFYY147 pKa = 10.81NDD149 pKa = 3.92EE150 pKa = 4.42SDD152 pKa = 2.95ITLQIGQEE160 pKa = 4.1EE161 pKa = 5.19YY162 pKa = 10.46IRR164 pKa = 11.84VYY166 pKa = 11.17NDD168 pKa = 2.86TGATLLNGTPVYY180 pKa = 9.92LTGEE184 pKa = 4.22SGATPTVSAARR195 pKa = 11.84ADD197 pKa = 3.56TTYY200 pKa = 11.4AEE202 pKa = 4.29SQAVGVTTHH211 pKa = 7.36DD212 pKa = 3.77IEE214 pKa = 4.83NNTIGYY220 pKa = 6.12VTVRR224 pKa = 11.84GLIADD229 pKa = 3.75VDD231 pKa = 4.26TSHH234 pKa = 6.66LTVGEE239 pKa = 4.17PVHH242 pKa = 5.99VAVGGGTVTEE252 pKa = 4.55SPSYY256 pKa = 9.94PYY258 pKa = 10.78YY259 pKa = 7.51PTEE262 pKa = 3.95VGVCLISAAIGGCIYY277 pKa = 10.91VSISSEE283 pKa = 4.05TFQTMRR289 pKa = 11.84VDD291 pKa = 3.88GNSHH295 pKa = 6.63FDD297 pKa = 3.47ADD299 pKa = 3.87VTIDD303 pKa = 3.41GDD305 pKa = 4.14LVVNGTQTITSSNNIALSGAFSYY328 pKa = 10.82FNSGDD333 pKa = 3.63TIGADD338 pKa = 3.05NTSFTGTGTNDD349 pKa = 3.16GVFTGHH355 pKa = 6.57YY356 pKa = 10.13NGTSSNKK363 pKa = 8.16TYY365 pKa = 10.48YY366 pKa = 10.65VRR368 pKa = 11.84ISDD371 pKa = 4.16DD372 pKa = 3.02QSAPDD377 pKa = 3.52EE378 pKa = 4.79FEE380 pKa = 3.74WSLDD384 pKa = 3.4NFVTIEE390 pKa = 3.91ATGIAITTDD399 pKa = 3.39DD400 pKa = 3.46QALEE404 pKa = 3.96EE405 pKa = 4.23GVNIKK410 pKa = 10.34FNADD414 pKa = 3.24NGHH417 pKa = 5.73VLGDD421 pKa = 3.7VWSGTASPTNVDD433 pKa = 2.98TGIASNRR440 pKa = 11.84NTGTSGVGYY449 pKa = 6.79THH451 pKa = 6.88VGMYY455 pKa = 10.47YY456 pKa = 10.36DD457 pKa = 3.52VSSNYY462 pKa = 7.02WTFFDD467 pKa = 4.74EE468 pKa = 4.6YY469 pKa = 10.87APEE472 pKa = 4.08PTGTIDD478 pKa = 3.15TSHH481 pKa = 7.0ASFSYY486 pKa = 11.27GDD488 pKa = 3.27IKK490 pKa = 11.15VNSVIGDD497 pKa = 3.52VVGNLTGIASSATQLANNRR516 pKa = 11.84NITLSGDD523 pKa = 3.56VTGTAVFNGGADD535 pKa = 3.66ANITATVVNDD545 pKa = 3.29SHH547 pKa = 6.31THH549 pKa = 4.64DD550 pKa = 3.24TRR552 pKa = 11.84YY553 pKa = 10.15VQLAGDD559 pKa = 4.01TMSGTLNVPTVDD571 pKa = 3.99FGDD574 pKa = 3.55WTITEE579 pKa = 4.57SGGSLYY585 pKa = 10.46FAYY588 pKa = 10.4QGTNKK593 pKa = 10.24LKK595 pKa = 10.69LDD597 pKa = 3.63TSGTLSVTNDD607 pKa = 2.9VQTDD611 pKa = 3.47QTII614 pKa = 3.13
Molecular weight: 64.61 kDa
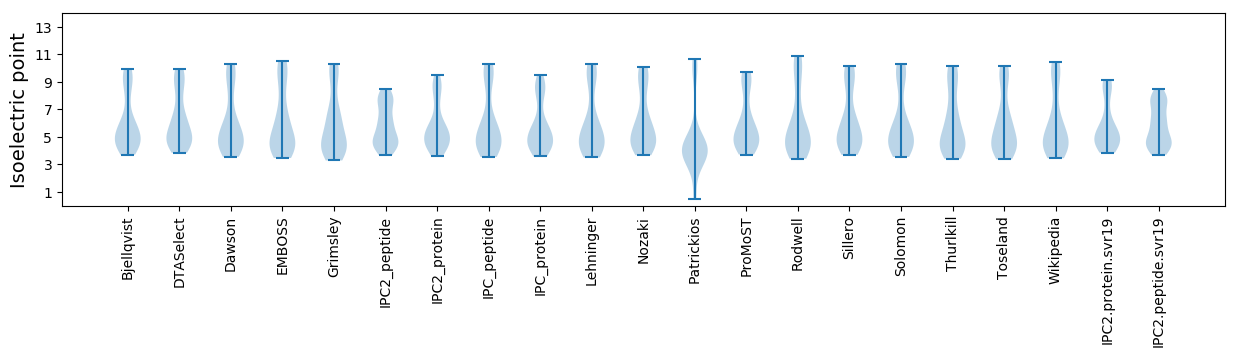
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A646QXS7|A0A646QXS7_9CAUD P4Hc domain-containing protein OS=Roseobacter phage CRP-6 OX=2559285 GN=CRP6_gp58 PE=4 SV=1
MM1 pKa = 8.5VDD3 pKa = 3.23MKK5 pKa = 11.35VNNSAYY11 pKa = 9.77KK12 pKa = 10.27KK13 pKa = 9.44LQSLEE18 pKa = 4.23EE19 pKa = 4.07YY20 pKa = 10.54AGQEE24 pKa = 4.57LEE26 pKa = 4.36DD27 pKa = 3.92KK28 pKa = 10.77LYY30 pKa = 11.0AIANDD35 pKa = 3.72AVRR38 pKa = 11.84TTLTSTSNKK47 pKa = 9.49GKK49 pKa = 10.26IGAVDD54 pKa = 3.44SGAYY58 pKa = 10.16LEE60 pKa = 4.75SFSFALGSGRR70 pKa = 11.84PRR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84VYY76 pKa = 10.56KK77 pKa = 10.64GSQKK81 pKa = 10.68SFNPEE86 pKa = 3.11QTALQNLLFDD96 pKa = 4.08IKK98 pKa = 10.81KK99 pKa = 9.57VDD101 pKa = 3.6LMSSTRR107 pKa = 11.84ITLRR111 pKa = 11.84NGSPYY116 pKa = 10.25ARR118 pKa = 11.84KK119 pKa = 9.53VEE121 pKa = 3.99YY122 pKa = 10.53NYY124 pKa = 10.76GYY126 pKa = 10.79RR127 pKa = 11.84IFAKK131 pKa = 10.23LRR133 pKa = 11.84RR134 pKa = 11.84KK135 pKa = 9.94HH136 pKa = 5.35GG137 pKa = 3.56
MM1 pKa = 8.5VDD3 pKa = 3.23MKK5 pKa = 11.35VNNSAYY11 pKa = 9.77KK12 pKa = 10.27KK13 pKa = 9.44LQSLEE18 pKa = 4.23EE19 pKa = 4.07YY20 pKa = 10.54AGQEE24 pKa = 4.57LEE26 pKa = 4.36DD27 pKa = 3.92KK28 pKa = 10.77LYY30 pKa = 11.0AIANDD35 pKa = 3.72AVRR38 pKa = 11.84TTLTSTSNKK47 pKa = 9.49GKK49 pKa = 10.26IGAVDD54 pKa = 3.44SGAYY58 pKa = 10.16LEE60 pKa = 4.75SFSFALGSGRR70 pKa = 11.84PRR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84VYY76 pKa = 10.56KK77 pKa = 10.64GSQKK81 pKa = 10.68SFNPEE86 pKa = 3.11QTALQNLLFDD96 pKa = 4.08IKK98 pKa = 10.81KK99 pKa = 9.57VDD101 pKa = 3.6LMSSTRR107 pKa = 11.84ITLRR111 pKa = 11.84NGSPYY116 pKa = 10.25ARR118 pKa = 11.84KK119 pKa = 9.53VEE121 pKa = 3.99YY122 pKa = 10.53NYY124 pKa = 10.76GYY126 pKa = 10.79RR127 pKa = 11.84IFAKK131 pKa = 10.23LRR133 pKa = 11.84RR134 pKa = 11.84KK135 pKa = 9.94HH136 pKa = 5.35GG137 pKa = 3.56
Molecular weight: 15.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14172 |
44 |
895 |
205.4 |
22.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.355 ± 0.422 | 0.946 ± 0.137 |
6.739 ± 0.284 | 6.548 ± 0.334 |
3.662 ± 0.177 | 7.988 ± 0.657 |
1.672 ± 0.143 | 5.151 ± 0.212 |
6.097 ± 0.388 | 7.663 ± 0.365 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.745 ± 0.205 | 4.558 ± 0.237 |
3.458 ± 0.182 | 3.359 ± 0.193 |
4.283 ± 0.285 | 6.809 ± 0.321 |
7.472 ± 0.566 | 7.226 ± 0.238 |
1.63 ± 0.138 | 3.641 ± 0.239 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
