
Maribacter cobaltidurans
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Maribacter
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
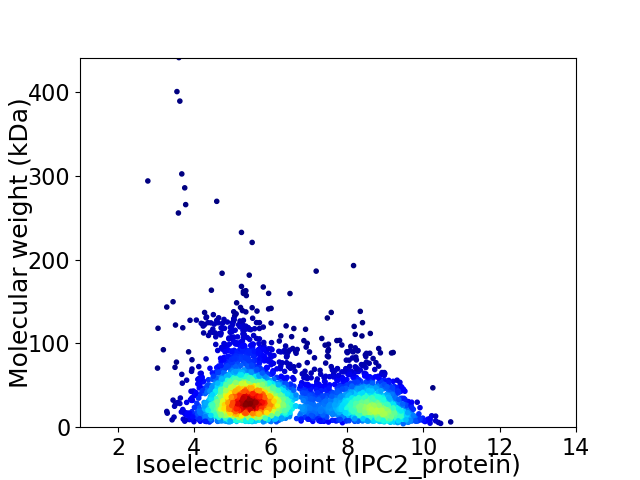
Virtual 2D-PAGE plot for 4004 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A223VA18|A0A223VA18_9FLAO OsmC family peroxiredoxin OS=Maribacter cobaltidurans OX=1178778 GN=CJ263_19445 PE=4 SV=1
MM1 pKa = 7.31AQMTFADD8 pKa = 4.4NFDD11 pKa = 3.49TSDD14 pKa = 3.62YY15 pKa = 11.33DD16 pKa = 3.71RR17 pKa = 11.84QDD19 pKa = 3.1GNTYY23 pKa = 9.36WATDD27 pKa = 4.04WIEE30 pKa = 4.48SDD32 pKa = 4.77DD33 pKa = 4.36SNLGPTSQYY42 pKa = 10.75IRR44 pKa = 11.84IQSQQLYY51 pKa = 8.76MYY53 pKa = 8.47YY54 pKa = 10.62LWTEE58 pKa = 4.67DD59 pKa = 2.97IRR61 pKa = 11.84RR62 pKa = 11.84TANLTGATSATLSFDD77 pKa = 3.34YY78 pKa = 7.9TTNNLGGTRR87 pKa = 11.84RR88 pKa = 11.84LGVYY92 pKa = 10.18ISSTGGAPYY101 pKa = 8.96TQIGTLSGNGTFTQDD116 pKa = 2.05ISAYY120 pKa = 9.13ISANTVIRR128 pKa = 11.84FAKK131 pKa = 10.53SNANWDD137 pKa = 4.01SNHH140 pKa = 6.06NASIDD145 pKa = 3.65NVLISAVTPVDD156 pKa = 3.27TDD158 pKa = 3.21GDD160 pKa = 4.33GIEE163 pKa = 5.48DD164 pKa = 4.57VADD167 pKa = 4.31LDD169 pKa = 4.05NDD171 pKa = 3.48NDD173 pKa = 5.9GILDD177 pKa = 3.54WDD179 pKa = 3.66EE180 pKa = 4.3CTFVATTTQNFNANGGTSVTNTSTSGRR207 pKa = 11.84GVMYY211 pKa = 10.22IDD213 pKa = 6.15FISIDD218 pKa = 3.31NSFNLTVNGTNIAAEE233 pKa = 4.31FQFQPGAPGNFARR246 pKa = 11.84FNTGFTYY253 pKa = 11.01GEE255 pKa = 4.47GGLPQLWSLTGTLANPVLRR274 pKa = 11.84VIVDD278 pKa = 3.55ADD280 pKa = 3.48GHH282 pKa = 6.38LKK284 pKa = 10.83LFGAQSSGGTLEE296 pKa = 4.4PLILDD301 pKa = 4.42TPPEE305 pKa = 4.23TVPWNPSGTNTITIGQFVTGPTNMNGQIRR334 pKa = 11.84FSGEE338 pKa = 3.77CDD340 pKa = 3.09TDD342 pKa = 3.45GDD344 pKa = 4.81GIFNRR349 pKa = 11.84FDD351 pKa = 4.25LDD353 pKa = 3.45SDD355 pKa = 3.62NDD357 pKa = 4.58GIYY360 pKa = 10.79DD361 pKa = 3.63AVEE364 pKa = 4.31AGHH367 pKa = 6.28GQAITNGAINGAVGTDD383 pKa = 3.6GVPNLVQTDD392 pKa = 3.97PNGGSINYY400 pKa = 9.74AYY402 pKa = 9.42TDD404 pKa = 3.49SDD406 pKa = 3.66GDD408 pKa = 4.28GILDD412 pKa = 4.65SIEE415 pKa = 4.76LDD417 pKa = 3.97SDD419 pKa = 3.73NDD421 pKa = 3.63GCNDD425 pKa = 3.14VLEE428 pKa = 5.58AGFTDD433 pKa = 3.68NNADD437 pKa = 3.56GFLGPNPVATDD448 pKa = 3.53GNGVVTSGTDD458 pKa = 3.31GYY460 pKa = 9.1TIPAEE465 pKa = 4.22GDD467 pKa = 3.11LDD469 pKa = 4.24SVFDD473 pKa = 3.9YY474 pKa = 10.5RR475 pKa = 11.84QAGAPPSINTQPVNTIICPGCNGSLSVTATNANLYY510 pKa = 8.28QWQRR514 pKa = 11.84FVGGSWANITDD525 pKa = 3.33NAMFSGANTATLNINNPTPSEE546 pKa = 3.81NGEE549 pKa = 4.01QYY551 pKa = 10.68RR552 pKa = 11.84VVLTNIAYY560 pKa = 10.03SCAQTTSATASLQIRR575 pKa = 11.84VNTIVTNRR583 pKa = 11.84RR584 pKa = 11.84ITYY587 pKa = 9.74RR588 pKa = 11.84VQKK591 pKa = 10.59NN592 pKa = 2.98
MM1 pKa = 7.31AQMTFADD8 pKa = 4.4NFDD11 pKa = 3.49TSDD14 pKa = 3.62YY15 pKa = 11.33DD16 pKa = 3.71RR17 pKa = 11.84QDD19 pKa = 3.1GNTYY23 pKa = 9.36WATDD27 pKa = 4.04WIEE30 pKa = 4.48SDD32 pKa = 4.77DD33 pKa = 4.36SNLGPTSQYY42 pKa = 10.75IRR44 pKa = 11.84IQSQQLYY51 pKa = 8.76MYY53 pKa = 8.47YY54 pKa = 10.62LWTEE58 pKa = 4.67DD59 pKa = 2.97IRR61 pKa = 11.84RR62 pKa = 11.84TANLTGATSATLSFDD77 pKa = 3.34YY78 pKa = 7.9TTNNLGGTRR87 pKa = 11.84RR88 pKa = 11.84LGVYY92 pKa = 10.18ISSTGGAPYY101 pKa = 8.96TQIGTLSGNGTFTQDD116 pKa = 2.05ISAYY120 pKa = 9.13ISANTVIRR128 pKa = 11.84FAKK131 pKa = 10.53SNANWDD137 pKa = 4.01SNHH140 pKa = 6.06NASIDD145 pKa = 3.65NVLISAVTPVDD156 pKa = 3.27TDD158 pKa = 3.21GDD160 pKa = 4.33GIEE163 pKa = 5.48DD164 pKa = 4.57VADD167 pKa = 4.31LDD169 pKa = 4.05NDD171 pKa = 3.48NDD173 pKa = 5.9GILDD177 pKa = 3.54WDD179 pKa = 3.66EE180 pKa = 4.3CTFVATTTQNFNANGGTSVTNTSTSGRR207 pKa = 11.84GVMYY211 pKa = 10.22IDD213 pKa = 6.15FISIDD218 pKa = 3.31NSFNLTVNGTNIAAEE233 pKa = 4.31FQFQPGAPGNFARR246 pKa = 11.84FNTGFTYY253 pKa = 11.01GEE255 pKa = 4.47GGLPQLWSLTGTLANPVLRR274 pKa = 11.84VIVDD278 pKa = 3.55ADD280 pKa = 3.48GHH282 pKa = 6.38LKK284 pKa = 10.83LFGAQSSGGTLEE296 pKa = 4.4PLILDD301 pKa = 4.42TPPEE305 pKa = 4.23TVPWNPSGTNTITIGQFVTGPTNMNGQIRR334 pKa = 11.84FSGEE338 pKa = 3.77CDD340 pKa = 3.09TDD342 pKa = 3.45GDD344 pKa = 4.81GIFNRR349 pKa = 11.84FDD351 pKa = 4.25LDD353 pKa = 3.45SDD355 pKa = 3.62NDD357 pKa = 4.58GIYY360 pKa = 10.79DD361 pKa = 3.63AVEE364 pKa = 4.31AGHH367 pKa = 6.28GQAITNGAINGAVGTDD383 pKa = 3.6GVPNLVQTDD392 pKa = 3.97PNGGSINYY400 pKa = 9.74AYY402 pKa = 9.42TDD404 pKa = 3.49SDD406 pKa = 3.66GDD408 pKa = 4.28GILDD412 pKa = 4.65SIEE415 pKa = 4.76LDD417 pKa = 3.97SDD419 pKa = 3.73NDD421 pKa = 3.63GCNDD425 pKa = 3.14VLEE428 pKa = 5.58AGFTDD433 pKa = 3.68NNADD437 pKa = 3.56GFLGPNPVATDD448 pKa = 3.53GNGVVTSGTDD458 pKa = 3.31GYY460 pKa = 9.1TIPAEE465 pKa = 4.22GDD467 pKa = 3.11LDD469 pKa = 4.24SVFDD473 pKa = 3.9YY474 pKa = 10.5RR475 pKa = 11.84QAGAPPSINTQPVNTIICPGCNGSLSVTATNANLYY510 pKa = 8.28QWQRR514 pKa = 11.84FVGGSWANITDD525 pKa = 3.33NAMFSGANTATLNINNPTPSEE546 pKa = 3.81NGEE549 pKa = 4.01QYY551 pKa = 10.68RR552 pKa = 11.84VVLTNIAYY560 pKa = 10.03SCAQTTSATASLQIRR575 pKa = 11.84VNTIVTNRR583 pKa = 11.84RR584 pKa = 11.84ITYY587 pKa = 9.74RR588 pKa = 11.84VQKK591 pKa = 10.59NN592 pKa = 2.98
Molecular weight: 63.06 kDa
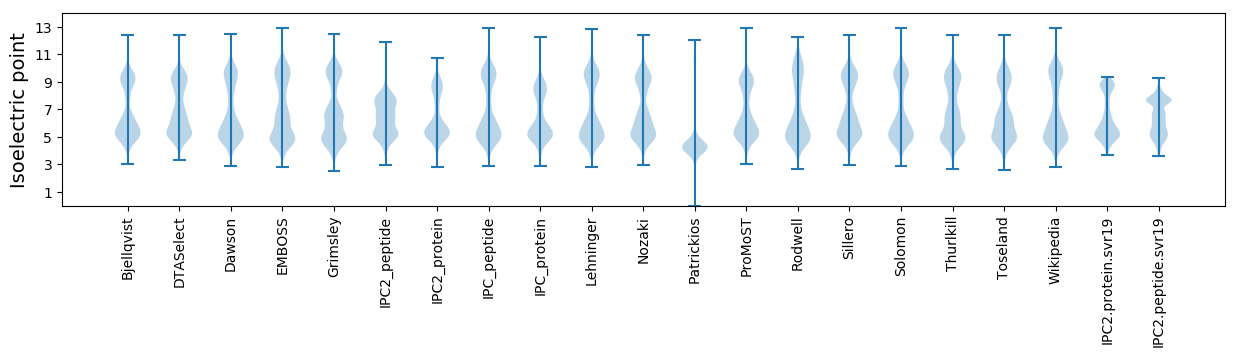
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A223V2Y0|A0A223V2Y0_9FLAO 50S ribosomal protein L25 OS=Maribacter cobaltidurans OX=1178778 GN=rplY PE=3 SV=1
MM1 pKa = 7.59QKK3 pKa = 9.07ATWLWRR9 pKa = 11.84VALTRR14 pKa = 11.84YY15 pKa = 9.58AGQEE19 pKa = 3.56LLGAIRR25 pKa = 11.84RR26 pKa = 11.84LFAIAASWEE35 pKa = 4.08KK36 pKa = 11.07CPFFQCGARR45 pKa = 11.84ANFGLFRR52 pKa = 11.84INIGNKK58 pKa = 8.37QSYY61 pKa = 9.49CVAFGGKK68 pKa = 9.12
MM1 pKa = 7.59QKK3 pKa = 9.07ATWLWRR9 pKa = 11.84VALTRR14 pKa = 11.84YY15 pKa = 9.58AGQEE19 pKa = 3.56LLGAIRR25 pKa = 11.84RR26 pKa = 11.84LFAIAASWEE35 pKa = 4.08KK36 pKa = 11.07CPFFQCGARR45 pKa = 11.84ANFGLFRR52 pKa = 11.84INIGNKK58 pKa = 8.37QSYY61 pKa = 9.49CVAFGGKK68 pKa = 9.12
Molecular weight: 7.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1370011 |
34 |
4164 |
342.2 |
38.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.21 ± 0.036 | 0.704 ± 0.013 |
5.758 ± 0.041 | 6.888 ± 0.048 |
5.157 ± 0.032 | 6.836 ± 0.048 |
1.789 ± 0.021 | 7.473 ± 0.036 |
7.412 ± 0.061 | 9.557 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.383 ± 0.021 | 5.816 ± 0.037 |
3.6 ± 0.027 | 3.355 ± 0.021 |
3.718 ± 0.026 | 6.43 ± 0.027 |
5.58 ± 0.053 | 6.212 ± 0.034 |
1.14 ± 0.016 | 3.983 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
