
Monosporascus sp. CRB-9-2
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Xylariomycetidae; Xylariales; Xylariales incertae sedis; Monosporascus; unclassified Monosporascus
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
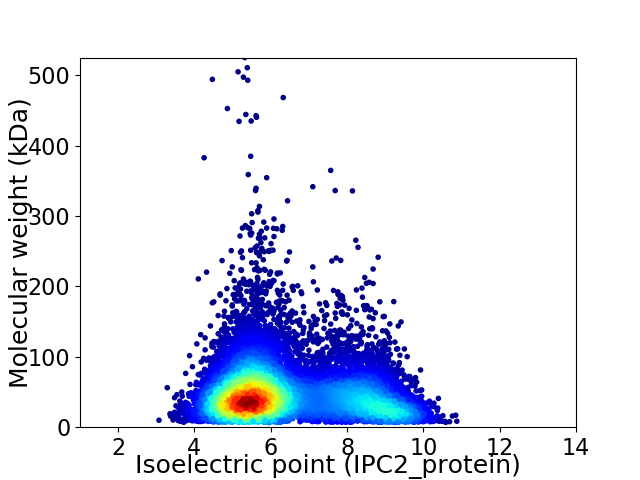
Virtual 2D-PAGE plot for 11997 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q4YHI6|A0A4Q4YHI6_9PEZI ABC1 domain-containing protein OS=Monosporascus sp. CRB-9-2 OX=2211643 GN=DL770_007698 PE=4 SV=1
MM1 pKa = 7.27NRR3 pKa = 11.84LLVLFTCLLAAGNASPFVPDD23 pKa = 3.92IKK25 pKa = 10.87PRR27 pKa = 11.84DD28 pKa = 3.55ICDD31 pKa = 3.33GNTATTRR38 pKa = 11.84TEE40 pKa = 3.42WCDD43 pKa = 3.17FDD45 pKa = 6.49INTDD49 pKa = 3.63YY50 pKa = 11.5YY51 pKa = 10.37NTVPEE56 pKa = 4.23TGVTRR61 pKa = 11.84EE62 pKa = 4.15YY63 pKa = 10.76FFEE66 pKa = 4.48LQDD69 pKa = 3.59VTVSPDD75 pKa = 3.1GVSRR79 pKa = 11.84SAIAVNGSIPGPTIFADD96 pKa = 3.05WGDD99 pKa = 3.8TVVVHH104 pKa = 5.46VTNSLTTSGNGSSIHH119 pKa = 5.28FHH121 pKa = 7.33GIRR124 pKa = 11.84QNFTNQNDD132 pKa = 4.22GVVSITQCPTAVGDD146 pKa = 3.54ATTYY150 pKa = 7.44TWRR153 pKa = 11.84ATQYY157 pKa = 10.72GSTWYY162 pKa = 9.69HH163 pKa = 4.67SHH165 pKa = 7.29FALQAWEE172 pKa = 4.54GVFGGIIINGPASSNYY188 pKa = 10.4DD189 pKa = 3.28EE190 pKa = 5.03DD191 pKa = 4.78LGMVFLNDD199 pKa = 3.43WSHH202 pKa = 4.29QTVDD206 pKa = 3.89EE207 pKa = 5.1LYY209 pKa = 7.78MTAQTSGPPTLDD221 pKa = 2.86NGLINGTNVFGSDD234 pKa = 4.83DD235 pKa = 4.24SDD237 pKa = 3.85DD238 pKa = 3.79QTGSRR243 pKa = 11.84LEE245 pKa = 4.01LSVTEE250 pKa = 4.13GTSYY254 pKa = 11.26RR255 pKa = 11.84LRR257 pKa = 11.84LVNAAIDD264 pKa = 3.54THH266 pKa = 6.65FKK268 pKa = 10.84FSVDD272 pKa = 2.97NHH274 pKa = 5.46TLTVIGMDD282 pKa = 3.86LVPIEE287 pKa = 4.83PYY289 pKa = 7.93TTEE292 pKa = 3.9FVNIGIADD300 pKa = 3.88QATVATDD307 pKa = 3.28FWMRR311 pKa = 11.84AIPQVACSDD320 pKa = 3.55NDD322 pKa = 3.8SADD325 pKa = 3.49NIMAIVHH332 pKa = 6.14YY333 pKa = 9.85GDD335 pKa = 3.59STATPSTTAFEE346 pKa = 4.49YY347 pKa = 10.64TDD349 pKa = 3.47SCDD352 pKa = 5.76DD353 pKa = 3.67EE354 pKa = 5.74DD355 pKa = 4.67LSKK358 pKa = 10.95LVPIVSKK365 pKa = 11.08AVGDD369 pKa = 4.1DD370 pKa = 3.91SNDD373 pKa = 3.44LLEE376 pKa = 4.48IATVGAVGNLFKK388 pKa = 10.64WFLNSTTMVVDD399 pKa = 3.56WADD402 pKa = 3.42PTLLQLSNNITTFDD416 pKa = 4.04DD417 pKa = 3.36SDD419 pKa = 4.05AVIEE423 pKa = 5.36LPDD426 pKa = 4.03ADD428 pKa = 3.28EE429 pKa = 3.91WVYY432 pKa = 11.28VVIQTTFAVPHH443 pKa = 7.1PIHH446 pKa = 6.48LHH448 pKa = 5.3GFDD451 pKa = 4.77FFVLAQGTGTFSSDD465 pKa = 3.06SVTLNTDD472 pKa = 3.01NPPRR476 pKa = 11.84RR477 pKa = 11.84DD478 pKa = 3.33TAMLPASGYY487 pKa = 10.47LVLAFEE493 pKa = 4.81TDD495 pKa = 3.92NPGAWLMHH503 pKa = 5.82CHH505 pKa = 6.47IGWHH509 pKa = 4.85TSEE512 pKa = 4.88GFALQFVVRR521 pKa = 11.84RR522 pKa = 11.84DD523 pKa = 3.8EE524 pKa = 4.26ILDD527 pKa = 4.87LIDD530 pKa = 3.63STTLEE535 pKa = 4.53DD536 pKa = 3.5TCSAWSSYY544 pKa = 11.3AEE546 pKa = 4.08EE547 pKa = 4.64SSVEE551 pKa = 3.95QDD553 pKa = 3.26DD554 pKa = 4.42SGVV557 pKa = 3.12
MM1 pKa = 7.27NRR3 pKa = 11.84LLVLFTCLLAAGNASPFVPDD23 pKa = 3.92IKK25 pKa = 10.87PRR27 pKa = 11.84DD28 pKa = 3.55ICDD31 pKa = 3.33GNTATTRR38 pKa = 11.84TEE40 pKa = 3.42WCDD43 pKa = 3.17FDD45 pKa = 6.49INTDD49 pKa = 3.63YY50 pKa = 11.5YY51 pKa = 10.37NTVPEE56 pKa = 4.23TGVTRR61 pKa = 11.84EE62 pKa = 4.15YY63 pKa = 10.76FFEE66 pKa = 4.48LQDD69 pKa = 3.59VTVSPDD75 pKa = 3.1GVSRR79 pKa = 11.84SAIAVNGSIPGPTIFADD96 pKa = 3.05WGDD99 pKa = 3.8TVVVHH104 pKa = 5.46VTNSLTTSGNGSSIHH119 pKa = 5.28FHH121 pKa = 7.33GIRR124 pKa = 11.84QNFTNQNDD132 pKa = 4.22GVVSITQCPTAVGDD146 pKa = 3.54ATTYY150 pKa = 7.44TWRR153 pKa = 11.84ATQYY157 pKa = 10.72GSTWYY162 pKa = 9.69HH163 pKa = 4.67SHH165 pKa = 7.29FALQAWEE172 pKa = 4.54GVFGGIIINGPASSNYY188 pKa = 10.4DD189 pKa = 3.28EE190 pKa = 5.03DD191 pKa = 4.78LGMVFLNDD199 pKa = 3.43WSHH202 pKa = 4.29QTVDD206 pKa = 3.89EE207 pKa = 5.1LYY209 pKa = 7.78MTAQTSGPPTLDD221 pKa = 2.86NGLINGTNVFGSDD234 pKa = 4.83DD235 pKa = 4.24SDD237 pKa = 3.85DD238 pKa = 3.79QTGSRR243 pKa = 11.84LEE245 pKa = 4.01LSVTEE250 pKa = 4.13GTSYY254 pKa = 11.26RR255 pKa = 11.84LRR257 pKa = 11.84LVNAAIDD264 pKa = 3.54THH266 pKa = 6.65FKK268 pKa = 10.84FSVDD272 pKa = 2.97NHH274 pKa = 5.46TLTVIGMDD282 pKa = 3.86LVPIEE287 pKa = 4.83PYY289 pKa = 7.93TTEE292 pKa = 3.9FVNIGIADD300 pKa = 3.88QATVATDD307 pKa = 3.28FWMRR311 pKa = 11.84AIPQVACSDD320 pKa = 3.55NDD322 pKa = 3.8SADD325 pKa = 3.49NIMAIVHH332 pKa = 6.14YY333 pKa = 9.85GDD335 pKa = 3.59STATPSTTAFEE346 pKa = 4.49YY347 pKa = 10.64TDD349 pKa = 3.47SCDD352 pKa = 5.76DD353 pKa = 3.67EE354 pKa = 5.74DD355 pKa = 4.67LSKK358 pKa = 10.95LVPIVSKK365 pKa = 11.08AVGDD369 pKa = 4.1DD370 pKa = 3.91SNDD373 pKa = 3.44LLEE376 pKa = 4.48IATVGAVGNLFKK388 pKa = 10.64WFLNSTTMVVDD399 pKa = 3.56WADD402 pKa = 3.42PTLLQLSNNITTFDD416 pKa = 4.04DD417 pKa = 3.36SDD419 pKa = 4.05AVIEE423 pKa = 5.36LPDD426 pKa = 4.03ADD428 pKa = 3.28EE429 pKa = 3.91WVYY432 pKa = 11.28VVIQTTFAVPHH443 pKa = 7.1PIHH446 pKa = 6.48LHH448 pKa = 5.3GFDD451 pKa = 4.77FFVLAQGTGTFSSDD465 pKa = 3.06SVTLNTDD472 pKa = 3.01NPPRR476 pKa = 11.84RR477 pKa = 11.84DD478 pKa = 3.33TAMLPASGYY487 pKa = 10.47LVLAFEE493 pKa = 4.81TDD495 pKa = 3.92NPGAWLMHH503 pKa = 5.82CHH505 pKa = 6.47IGWHH509 pKa = 4.85TSEE512 pKa = 4.88GFALQFVVRR521 pKa = 11.84RR522 pKa = 11.84DD523 pKa = 3.8EE524 pKa = 4.26ILDD527 pKa = 4.87LIDD530 pKa = 3.63STTLEE535 pKa = 4.53DD536 pKa = 3.5TCSAWSSYY544 pKa = 11.3AEE546 pKa = 4.08EE547 pKa = 4.64SSVEE551 pKa = 3.95QDD553 pKa = 3.26DD554 pKa = 4.42SGVV557 pKa = 3.12
Molecular weight: 60.83 kDa
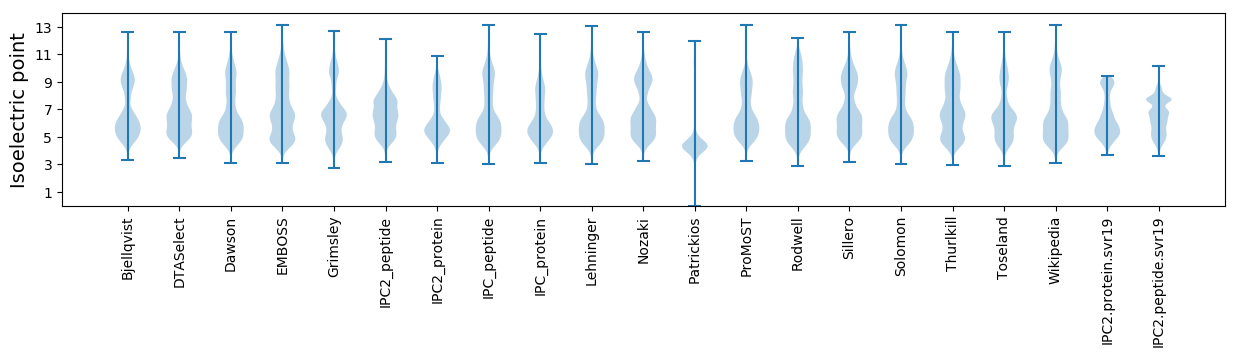
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q4WBM7|A0A4Q4WBM7_9PEZI Uncharacterized protein OS=Monosporascus sp. CRB-9-2 OX=2211643 GN=DL770_011809 PE=4 SV=1
MM1 pKa = 7.46RR2 pKa = 11.84RR3 pKa = 11.84ARR5 pKa = 11.84SRR7 pKa = 11.84KK8 pKa = 7.05TSPSGVRR15 pKa = 11.84LMRR18 pKa = 11.84RR19 pKa = 11.84VVRR22 pKa = 11.84VTRR25 pKa = 11.84GMPMRR30 pKa = 11.84ASICARR36 pKa = 11.84RR37 pKa = 11.84LLTAGVVMPRR47 pKa = 11.84LRR49 pKa = 11.84AAALRR54 pKa = 11.84LPAVASATKK63 pKa = 9.84KK64 pKa = 10.09PSSAEE69 pKa = 4.01PISNRR74 pKa = 11.84VHH76 pKa = 6.44AA77 pKa = 5.21
MM1 pKa = 7.46RR2 pKa = 11.84RR3 pKa = 11.84ARR5 pKa = 11.84SRR7 pKa = 11.84KK8 pKa = 7.05TSPSGVRR15 pKa = 11.84LMRR18 pKa = 11.84RR19 pKa = 11.84VVRR22 pKa = 11.84VTRR25 pKa = 11.84GMPMRR30 pKa = 11.84ASICARR36 pKa = 11.84RR37 pKa = 11.84LLTAGVVMPRR47 pKa = 11.84LRR49 pKa = 11.84AAALRR54 pKa = 11.84LPAVASATKK63 pKa = 9.84KK64 pKa = 10.09PSSAEE69 pKa = 4.01PISNRR74 pKa = 11.84VHH76 pKa = 6.44AA77 pKa = 5.21
Molecular weight: 8.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5711890 |
66 |
4747 |
476.1 |
52.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.294 ± 0.021 | 1.17 ± 0.008 |
5.907 ± 0.014 | 6.359 ± 0.022 |
3.537 ± 0.014 | 7.306 ± 0.022 |
2.334 ± 0.01 | 4.503 ± 0.014 |
4.586 ± 0.02 | 8.754 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.108 ± 0.008 | 3.508 ± 0.012 |
6.205 ± 0.026 | 3.873 ± 0.017 |
6.592 ± 0.022 | 7.815 ± 0.021 |
5.771 ± 0.013 | 6.202 ± 0.015 |
1.465 ± 0.008 | 2.709 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
