
Rice tungro bacilliform virus (isolate Philippines) (RTBV)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Tungrovirus; Rice tungro bacilliform virus
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
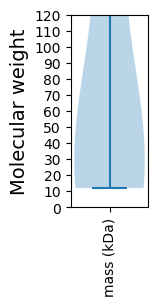
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>sp|P27502|POL_RTBVP Polyprotein P3 OS=Rice tungro bacilliform virus (isolate Philippines) OX=10655 PE=1 SV=1
MM1 pKa = 7.34NIEE4 pKa = 4.18YY5 pKa = 8.91PYY7 pKa = 10.17SIHH10 pKa = 8.09IIDD13 pKa = 4.04KK14 pKa = 10.13NKK16 pKa = 9.96VPIYY20 pKa = 10.09DD21 pKa = 3.47QGNLFHH27 pKa = 6.52TEE29 pKa = 3.4KK30 pKa = 10.56SSRR33 pKa = 11.84LSHH36 pKa = 6.28VSRR39 pKa = 11.84GLLDD43 pKa = 3.45HH44 pKa = 7.03LFTFSSDD51 pKa = 2.72NTEE54 pKa = 3.94RR55 pKa = 11.84VRR57 pKa = 11.84KK58 pKa = 9.12LHH60 pKa = 6.43ILADD64 pKa = 3.74YY65 pKa = 10.82LYY67 pKa = 9.83LLEE70 pKa = 4.98SEE72 pKa = 4.29RR73 pKa = 11.84EE74 pKa = 4.14SYY76 pKa = 10.09KK77 pKa = 10.97NEE79 pKa = 3.94WISLKK84 pKa = 10.84DD85 pKa = 3.63QVSLLQKK92 pKa = 10.48QNSEE96 pKa = 3.56LRR98 pKa = 11.84ARR100 pKa = 11.84IATNKK105 pKa = 10.05EE106 pKa = 3.64IIEE109 pKa = 4.15GLRR112 pKa = 11.84EE113 pKa = 3.97PVKK116 pKa = 10.83KK117 pKa = 10.14PIYY120 pKa = 6.32TTQDD124 pKa = 2.89KK125 pKa = 10.68EE126 pKa = 3.92RR127 pKa = 11.84LRR129 pKa = 11.84VFFCEE134 pKa = 4.12EE135 pKa = 3.35RR136 pKa = 11.84SMEE139 pKa = 4.06YY140 pKa = 9.57IYY142 pKa = 11.1YY143 pKa = 9.86HH144 pKa = 6.91IKK146 pKa = 10.55RR147 pKa = 11.84LAQQSYY153 pKa = 10.0YY154 pKa = 11.22SHH156 pKa = 7.29LNNLQKK162 pKa = 10.8DD163 pKa = 4.07CEE165 pKa = 4.34PFRR168 pKa = 11.84GVYY171 pKa = 9.44MSFLTNVKK179 pKa = 10.18FLVLCEE185 pKa = 3.89AGYY188 pKa = 8.13WTVPDD193 pKa = 3.8IEE195 pKa = 4.8TNTTEE200 pKa = 4.7SILSLSQKK208 pKa = 10.29KK209 pKa = 10.51GEE211 pKa = 4.48DD212 pKa = 3.48LLQKK216 pKa = 10.69GVVIFNEE223 pKa = 4.23LEE225 pKa = 3.86GGYY228 pKa = 9.33QLSPRR233 pKa = 11.84FIGDD237 pKa = 3.93LYY239 pKa = 11.01AHH241 pKa = 6.92GFIKK245 pKa = 10.42QINFTTKK252 pKa = 9.93VPEE255 pKa = 4.18GLPPIIAEE263 pKa = 4.21KK264 pKa = 10.02LQDD267 pKa = 3.65YY268 pKa = 9.46KK269 pKa = 11.37FPGSNTVLIEE279 pKa = 4.08RR280 pKa = 11.84EE281 pKa = 3.73IPRR284 pKa = 11.84WNFNEE289 pKa = 4.38MKK291 pKa = 10.28RR292 pKa = 11.84EE293 pKa = 3.88TQMRR297 pKa = 11.84TNLYY301 pKa = 8.54IFKK304 pKa = 10.31NYY306 pKa = 9.86RR307 pKa = 11.84CFYY310 pKa = 9.51GYY312 pKa = 10.71SPLRR316 pKa = 11.84PYY318 pKa = 10.91EE319 pKa = 4.46PITPEE324 pKa = 3.74EE325 pKa = 4.15FGFDD329 pKa = 3.61YY330 pKa = 11.02YY331 pKa = 11.27SWEE334 pKa = 3.98NMVDD338 pKa = 3.45EE339 pKa = 5.32DD340 pKa = 4.04EE341 pKa = 5.46GEE343 pKa = 4.06VVYY346 pKa = 10.58ISKK349 pKa = 8.2YY350 pKa = 7.64TKK352 pKa = 9.64IIKK355 pKa = 7.73VTKK358 pKa = 7.54EE359 pKa = 3.94HH360 pKa = 5.6AWAWPEE366 pKa = 3.77HH367 pKa = 6.98DD368 pKa = 5.06GDD370 pKa = 4.27TMSCTTSIEE379 pKa = 4.24DD380 pKa = 3.47EE381 pKa = 4.55WIHH384 pKa = 7.91RR385 pKa = 11.84MDD387 pKa = 3.61NAA389 pKa = 4.25
MM1 pKa = 7.34NIEE4 pKa = 4.18YY5 pKa = 8.91PYY7 pKa = 10.17SIHH10 pKa = 8.09IIDD13 pKa = 4.04KK14 pKa = 10.13NKK16 pKa = 9.96VPIYY20 pKa = 10.09DD21 pKa = 3.47QGNLFHH27 pKa = 6.52TEE29 pKa = 3.4KK30 pKa = 10.56SSRR33 pKa = 11.84LSHH36 pKa = 6.28VSRR39 pKa = 11.84GLLDD43 pKa = 3.45HH44 pKa = 7.03LFTFSSDD51 pKa = 2.72NTEE54 pKa = 3.94RR55 pKa = 11.84VRR57 pKa = 11.84KK58 pKa = 9.12LHH60 pKa = 6.43ILADD64 pKa = 3.74YY65 pKa = 10.82LYY67 pKa = 9.83LLEE70 pKa = 4.98SEE72 pKa = 4.29RR73 pKa = 11.84EE74 pKa = 4.14SYY76 pKa = 10.09KK77 pKa = 10.97NEE79 pKa = 3.94WISLKK84 pKa = 10.84DD85 pKa = 3.63QVSLLQKK92 pKa = 10.48QNSEE96 pKa = 3.56LRR98 pKa = 11.84ARR100 pKa = 11.84IATNKK105 pKa = 10.05EE106 pKa = 3.64IIEE109 pKa = 4.15GLRR112 pKa = 11.84EE113 pKa = 3.97PVKK116 pKa = 10.83KK117 pKa = 10.14PIYY120 pKa = 6.32TTQDD124 pKa = 2.89KK125 pKa = 10.68EE126 pKa = 3.92RR127 pKa = 11.84LRR129 pKa = 11.84VFFCEE134 pKa = 4.12EE135 pKa = 3.35RR136 pKa = 11.84SMEE139 pKa = 4.06YY140 pKa = 9.57IYY142 pKa = 11.1YY143 pKa = 9.86HH144 pKa = 6.91IKK146 pKa = 10.55RR147 pKa = 11.84LAQQSYY153 pKa = 10.0YY154 pKa = 11.22SHH156 pKa = 7.29LNNLQKK162 pKa = 10.8DD163 pKa = 4.07CEE165 pKa = 4.34PFRR168 pKa = 11.84GVYY171 pKa = 9.44MSFLTNVKK179 pKa = 10.18FLVLCEE185 pKa = 3.89AGYY188 pKa = 8.13WTVPDD193 pKa = 3.8IEE195 pKa = 4.8TNTTEE200 pKa = 4.7SILSLSQKK208 pKa = 10.29KK209 pKa = 10.51GEE211 pKa = 4.48DD212 pKa = 3.48LLQKK216 pKa = 10.69GVVIFNEE223 pKa = 4.23LEE225 pKa = 3.86GGYY228 pKa = 9.33QLSPRR233 pKa = 11.84FIGDD237 pKa = 3.93LYY239 pKa = 11.01AHH241 pKa = 6.92GFIKK245 pKa = 10.42QINFTTKK252 pKa = 9.93VPEE255 pKa = 4.18GLPPIIAEE263 pKa = 4.21KK264 pKa = 10.02LQDD267 pKa = 3.65YY268 pKa = 9.46KK269 pKa = 11.37FPGSNTVLIEE279 pKa = 4.08RR280 pKa = 11.84EE281 pKa = 3.73IPRR284 pKa = 11.84WNFNEE289 pKa = 4.38MKK291 pKa = 10.28RR292 pKa = 11.84EE293 pKa = 3.88TQMRR297 pKa = 11.84TNLYY301 pKa = 8.54IFKK304 pKa = 10.31NYY306 pKa = 9.86RR307 pKa = 11.84CFYY310 pKa = 9.51GYY312 pKa = 10.71SPLRR316 pKa = 11.84PYY318 pKa = 10.91EE319 pKa = 4.46PITPEE324 pKa = 3.74EE325 pKa = 4.15FGFDD329 pKa = 3.61YY330 pKa = 11.02YY331 pKa = 11.27SWEE334 pKa = 3.98NMVDD338 pKa = 3.45EE339 pKa = 5.32DD340 pKa = 4.04EE341 pKa = 5.46GEE343 pKa = 4.06VVYY346 pKa = 10.58ISKK349 pKa = 8.2YY350 pKa = 7.64TKK352 pKa = 9.64IIKK355 pKa = 7.73VTKK358 pKa = 7.54EE359 pKa = 3.94HH360 pKa = 5.6AWAWPEE366 pKa = 3.77HH367 pKa = 6.98DD368 pKa = 5.06GDD370 pKa = 4.27TMSCTTSIEE379 pKa = 4.24DD380 pKa = 3.47EE381 pKa = 4.55WIHH384 pKa = 7.91RR385 pKa = 11.84MDD387 pKa = 3.61NAA389 pKa = 4.25
Molecular weight: 46.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P27500|P1_RTBVP Protein P1 OS=Rice tungro bacilliform virus (isolate Philippines) OX=10655 PE=4 SV=3
MM1 pKa = 7.17SADD4 pKa = 3.3YY5 pKa = 8.21PTFKK9 pKa = 10.54EE10 pKa = 3.97ALEE13 pKa = 4.08KK14 pKa = 10.77FKK16 pKa = 11.18NLEE19 pKa = 3.71SDD21 pKa = 3.44TAGKK25 pKa = 10.42DD26 pKa = 2.97KK27 pKa = 10.96FNWVFTLEE35 pKa = 4.38NIKK38 pKa = 10.61SAADD42 pKa = 3.19VNLASKK48 pKa = 10.95GLVQLYY54 pKa = 10.22ALQEE58 pKa = 3.72IDD60 pKa = 4.56KK61 pKa = 10.49KK62 pKa = 11.09INNLTTQVSKK72 pKa = 11.28LPTTSGSSSAGAIVPAGSNTQGQYY96 pKa = 10.41KK97 pKa = 10.25APPKK101 pKa = 10.32KK102 pKa = 10.34GIKK105 pKa = 9.46RR106 pKa = 11.84KK107 pKa = 10.13YY108 pKa = 8.2PAA110 pKa = 4.41
MM1 pKa = 7.17SADD4 pKa = 3.3YY5 pKa = 8.21PTFKK9 pKa = 10.54EE10 pKa = 3.97ALEE13 pKa = 4.08KK14 pKa = 10.77FKK16 pKa = 11.18NLEE19 pKa = 3.71SDD21 pKa = 3.44TAGKK25 pKa = 10.42DD26 pKa = 2.97KK27 pKa = 10.96FNWVFTLEE35 pKa = 4.38NIKK38 pKa = 10.61SAADD42 pKa = 3.19VNLASKK48 pKa = 10.95GLVQLYY54 pKa = 10.22ALQEE58 pKa = 3.72IDD60 pKa = 4.56KK61 pKa = 10.49KK62 pKa = 11.09INNLTTQVSKK72 pKa = 11.28LPTTSGSSSAGAIVPAGSNTQGQYY96 pKa = 10.41KK97 pKa = 10.25APPKK101 pKa = 10.32KK102 pKa = 10.34GIKK105 pKa = 9.46RR106 pKa = 11.84KK107 pKa = 10.13YY108 pKa = 8.2PAA110 pKa = 4.41
Molecular weight: 11.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2373 |
110 |
1675 |
593.3 |
68.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.425 ± 1.059 | 1.391 ± 0.392 |
5.942 ± 0.514 | 9.313 ± 0.812 |
3.961 ± 0.221 | 4.256 ± 0.85 |
2.023 ± 0.379 | 8.386 ± 0.763 |
10.619 ± 1.651 | 8.091 ± 1.188 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.812 ± 0.523 | 5.9 ± 0.272 |
3.456 ± 0.547 | 3.751 ± 0.754 |
4.34 ± 0.684 | 6.49 ± 0.611 |
5.9 ± 0.207 | 4.214 ± 0.199 |
1.264 ± 0.255 | 4.467 ± 0.781 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
