
Echinococcus multilocularis (Fox tapeworm)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Platyhelminthes; Cestoda; Eucestoda; Cyclophyllidea; Taeniidae; Echinococcus
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
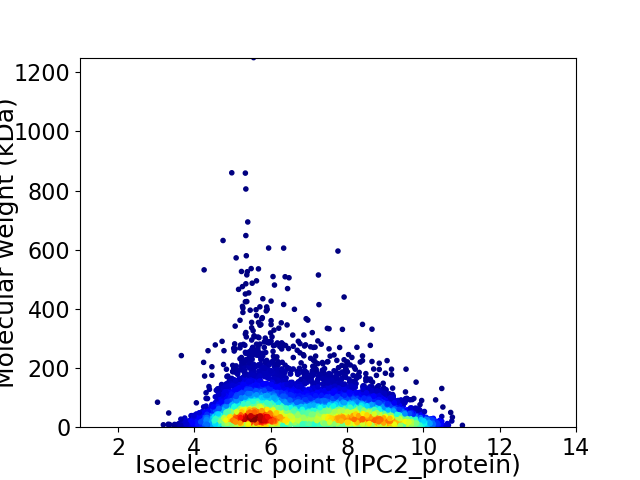
Virtual 2D-PAGE plot for 10472 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
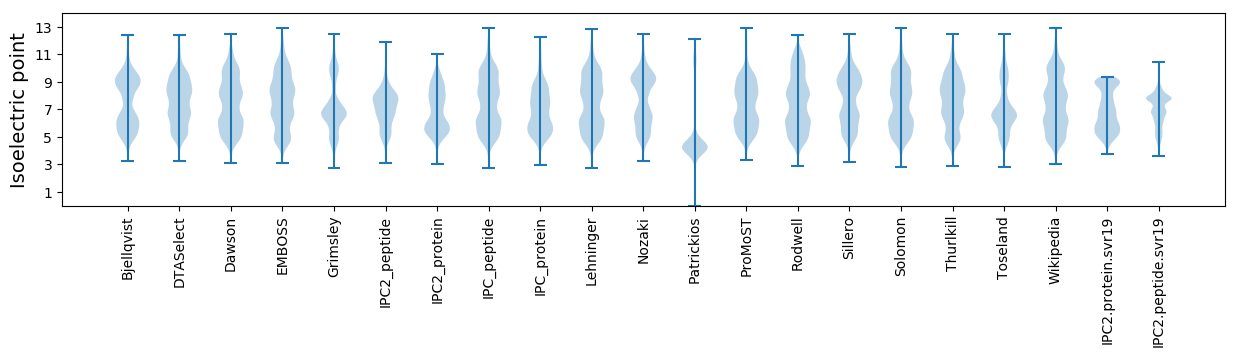
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A068XVC5|A0A068XVC5_ECHMU Forkhead box protein P4 OS=Echinococcus multilocularis OX=6211 GN=EmuJ_000320300 PE=4 SV=1
MM1 pKa = 7.54EE2 pKa = 6.48FIHH5 pKa = 6.83CEE7 pKa = 3.87KK8 pKa = 10.82NPTSDD13 pKa = 3.28VVKK16 pKa = 10.65SLEE19 pKa = 3.58EE20 pKa = 4.09LYY22 pKa = 10.66RR23 pKa = 11.84VEE25 pKa = 4.21TLVQIPEE32 pKa = 4.17WDD34 pKa = 3.58DD35 pKa = 2.96MGYY38 pKa = 10.75EE39 pKa = 4.3DD40 pKa = 5.08WHH42 pKa = 7.25ACNQPPQDD50 pKa = 4.38PNCEE54 pKa = 4.39VIPDD58 pKa = 3.67IAVDD62 pKa = 3.85ANVDD66 pKa = 4.38SIFGDD71 pKa = 3.53
MM1 pKa = 7.54EE2 pKa = 6.48FIHH5 pKa = 6.83CEE7 pKa = 3.87KK8 pKa = 10.82NPTSDD13 pKa = 3.28VVKK16 pKa = 10.65SLEE19 pKa = 3.58EE20 pKa = 4.09LYY22 pKa = 10.66RR23 pKa = 11.84VEE25 pKa = 4.21TLVQIPEE32 pKa = 4.17WDD34 pKa = 3.58DD35 pKa = 2.96MGYY38 pKa = 10.75EE39 pKa = 4.3DD40 pKa = 5.08WHH42 pKa = 7.25ACNQPPQDD50 pKa = 4.38PNCEE54 pKa = 4.39VIPDD58 pKa = 3.67IAVDD62 pKa = 3.85ANVDD66 pKa = 4.38SIFGDD71 pKa = 3.53
Molecular weight: 8.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A087VYH0|A0A087VYH0_ECHMU Metaxin 1 OS=Echinococcus multilocularis OX=6211 GN=EmuJ_000092500 PE=3 SV=1
MM1 pKa = 7.66LGQLTRR7 pKa = 11.84RR8 pKa = 11.84HH9 pKa = 5.71HH10 pKa = 6.63RR11 pKa = 11.84PRR13 pKa = 11.84DD14 pKa = 3.8TPHH17 pKa = 6.5EE18 pKa = 4.2ACAADD23 pKa = 4.19RR24 pKa = 11.84FLHH27 pKa = 6.81ASTGKK32 pKa = 10.12NSNTISRR39 pKa = 11.84DD40 pKa = 3.35MIGVQRR46 pKa = 11.84LMLFIAILRR55 pKa = 11.84VFVLPTIHH63 pKa = 6.2FTRR66 pKa = 11.84SASLLLQFILAVFALFVLHH85 pKa = 6.44YY86 pKa = 10.6LSLLLSTHH94 pKa = 5.45VEE96 pKa = 4.22RR97 pKa = 11.84PKK99 pKa = 10.39HH100 pKa = 5.67RR101 pKa = 11.84EE102 pKa = 3.4RR103 pKa = 11.84KK104 pKa = 9.31EE105 pKa = 3.71FMEE108 pKa = 4.07MKK110 pKa = 9.91VRR112 pKa = 11.84PEE114 pKa = 4.18SLPSPPPACFSHH126 pKa = 8.45LDD128 pKa = 3.31ILRR131 pKa = 11.84SQIHH135 pKa = 6.75LSPFSSSSSCVILRR149 pKa = 11.84QCHH152 pKa = 5.18FAHH155 pKa = 6.8PRR157 pKa = 11.84PHH159 pKa = 7.31LFTLSSQFTPEE170 pKa = 4.57SEE172 pKa = 4.2DD173 pKa = 3.15SLYY176 pKa = 11.17ASSIAEE182 pKa = 3.85TDD184 pKa = 2.99SRR186 pKa = 11.84GGIKK190 pKa = 10.05RR191 pKa = 11.84GRR193 pKa = 11.84KK194 pKa = 9.11AGFVADD200 pKa = 3.87AVTPVISGNYY210 pKa = 8.85CSSPPKK216 pKa = 10.27LRR218 pKa = 11.84LNHH221 pKa = 6.81PYY223 pKa = 10.5SLSSGVNNHH232 pKa = 5.67GQVQRR237 pKa = 11.84EE238 pKa = 3.86NQRR241 pKa = 11.84RR242 pKa = 11.84NRR244 pKa = 11.84VIKK247 pKa = 10.54RR248 pKa = 11.84NEE250 pKa = 4.01GVVKK254 pKa = 10.7SS255 pKa = 3.91
MM1 pKa = 7.66LGQLTRR7 pKa = 11.84RR8 pKa = 11.84HH9 pKa = 5.71HH10 pKa = 6.63RR11 pKa = 11.84PRR13 pKa = 11.84DD14 pKa = 3.8TPHH17 pKa = 6.5EE18 pKa = 4.2ACAADD23 pKa = 4.19RR24 pKa = 11.84FLHH27 pKa = 6.81ASTGKK32 pKa = 10.12NSNTISRR39 pKa = 11.84DD40 pKa = 3.35MIGVQRR46 pKa = 11.84LMLFIAILRR55 pKa = 11.84VFVLPTIHH63 pKa = 6.2FTRR66 pKa = 11.84SASLLLQFILAVFALFVLHH85 pKa = 6.44YY86 pKa = 10.6LSLLLSTHH94 pKa = 5.45VEE96 pKa = 4.22RR97 pKa = 11.84PKK99 pKa = 10.39HH100 pKa = 5.67RR101 pKa = 11.84EE102 pKa = 3.4RR103 pKa = 11.84KK104 pKa = 9.31EE105 pKa = 3.71FMEE108 pKa = 4.07MKK110 pKa = 9.91VRR112 pKa = 11.84PEE114 pKa = 4.18SLPSPPPACFSHH126 pKa = 8.45LDD128 pKa = 3.31ILRR131 pKa = 11.84SQIHH135 pKa = 6.75LSPFSSSSSCVILRR149 pKa = 11.84QCHH152 pKa = 5.18FAHH155 pKa = 6.8PRR157 pKa = 11.84PHH159 pKa = 7.31LFTLSSQFTPEE170 pKa = 4.57SEE172 pKa = 4.2DD173 pKa = 3.15SLYY176 pKa = 11.17ASSIAEE182 pKa = 3.85TDD184 pKa = 2.99SRR186 pKa = 11.84GGIKK190 pKa = 10.05RR191 pKa = 11.84GRR193 pKa = 11.84KK194 pKa = 9.11AGFVADD200 pKa = 3.87AVTPVISGNYY210 pKa = 8.85CSSPPKK216 pKa = 10.27LRR218 pKa = 11.84LNHH221 pKa = 6.81PYY223 pKa = 10.5SLSSGVNNHH232 pKa = 5.67GQVQRR237 pKa = 11.84EE238 pKa = 3.86NQRR241 pKa = 11.84RR242 pKa = 11.84NRR244 pKa = 11.84VIKK247 pKa = 10.54RR248 pKa = 11.84NEE250 pKa = 4.01GVVKK254 pKa = 10.7SS255 pKa = 3.91
Molecular weight: 28.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5200134 |
8 |
11193 |
496.6 |
55.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.479 ± 0.023 | 2.154 ± 0.017 |
4.968 ± 0.018 | 6.204 ± 0.028 |
3.952 ± 0.015 | 5.79 ± 0.027 |
2.527 ± 0.012 | 4.852 ± 0.016 |
4.757 ± 0.024 | 9.827 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.118 ± 0.01 | 3.99 ± 0.013 |
5.849 ± 0.028 | 4.006 ± 0.018 |
6.286 ± 0.018 | 9.169 ± 0.033 |
5.964 ± 0.022 | 6.381 ± 0.017 |
1.099 ± 0.007 | 2.628 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
