
Diachasmimorpha longicaudata rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 7.12
Get precalculated fractions of proteins
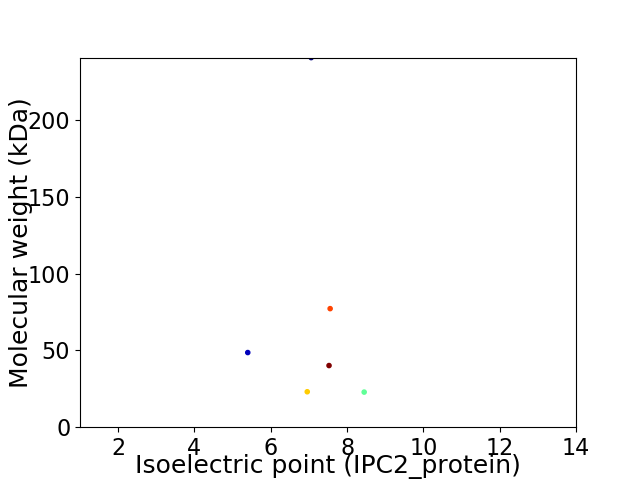
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0X9K747|A0A0X9K747_9RHAB Putative glycoprotein B OS=Diachasmimorpha longicaudata rhabdovirus OX=1585246 GN=G PE=4 SV=1
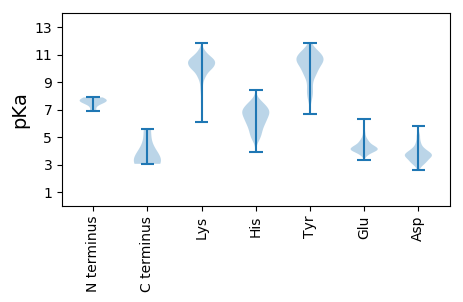
MM1 pKa = 7.57EE2 pKa = 6.3LYY4 pKa = 9.21NTIINGAKK12 pKa = 9.89EE13 pKa = 3.61IDD15 pKa = 3.45IGGSNPMTGALSGWTDD31 pKa = 3.38DD32 pKa = 5.05SYY34 pKa = 11.73KK35 pKa = 10.41QLYY38 pKa = 7.86GHH40 pKa = 6.46VRR42 pKa = 11.84ITQLEE47 pKa = 4.31VSFIPRR53 pKa = 11.84ILKK56 pKa = 9.9AWFSTYY62 pKa = 10.54YY63 pKa = 9.65NHH65 pKa = 7.61NIGNIDD71 pKa = 3.97LSSLMMSTLSLITVSSVSKK90 pKa = 10.38GQTSQTPNIQEE101 pKa = 4.17MISIAEE107 pKa = 3.94NLGKK111 pKa = 10.3VEE113 pKa = 3.99SQLRR117 pKa = 11.84KK118 pKa = 9.53NSPLIDD124 pKa = 4.01HH125 pKa = 7.39DD126 pKa = 4.53SEE128 pKa = 5.86KK129 pKa = 11.13EE130 pKa = 3.85DD131 pKa = 3.68ILAGDD136 pKa = 4.26HH137 pKa = 6.58EE138 pKa = 4.84EE139 pKa = 3.93QHH141 pKa = 6.96SIDD144 pKa = 4.17SSDD147 pKa = 3.36VMYY150 pKa = 11.07LSMISDD156 pKa = 4.36PDD158 pKa = 3.79YY159 pKa = 11.24QDD161 pKa = 3.12TLINIASFQALVMLRR176 pKa = 11.84HH177 pKa = 5.16ITKK180 pKa = 9.92TNTSVDD186 pKa = 3.06KK187 pKa = 10.58YY188 pKa = 10.55QRR190 pKa = 11.84EE191 pKa = 4.31SLTRR195 pKa = 11.84TLKK198 pKa = 10.64SLCPNTLLTADD209 pKa = 4.63IPPPSAQFVTYY220 pKa = 9.95FKK222 pKa = 10.6LNCMKK227 pKa = 10.7GNDD230 pKa = 3.55VATSLLAYY238 pKa = 10.38VMNLYY243 pKa = 9.53MEE245 pKa = 5.13ARR247 pKa = 11.84QSGATPLVDD256 pKa = 3.86YY257 pKa = 11.24LNAGCLTHH265 pKa = 7.25LKK267 pKa = 10.86GNGLALLEE275 pKa = 5.51LIFQVHH281 pKa = 6.89LCTKK285 pKa = 10.44LKK287 pKa = 10.87LSDD290 pKa = 4.47LFDD293 pKa = 3.47MTYY296 pKa = 10.88SSQCAQSIDD305 pKa = 3.29RR306 pKa = 11.84AYY308 pKa = 11.02HH309 pKa = 5.48HH310 pKa = 6.52FKK312 pKa = 10.57EE313 pKa = 4.22YY314 pKa = 11.4AEE316 pKa = 4.6LKK318 pKa = 10.84SNDD321 pKa = 3.17GKK323 pKa = 10.64IIRR326 pKa = 11.84PAQVTWWWARR336 pKa = 11.84LFNDD340 pKa = 4.37KK341 pKa = 11.04YY342 pKa = 11.15FMDD345 pKa = 3.58MSIRR349 pKa = 11.84NNYY352 pKa = 8.21QYY354 pKa = 11.04CLRR357 pKa = 11.84LACILTCYY365 pKa = 8.33QQQGSPLEE373 pKa = 4.01AAGFGTVSAKK383 pKa = 10.16EE384 pKa = 3.93LSKK387 pKa = 11.42HH388 pKa = 5.91NDD390 pKa = 2.9FATKK394 pKa = 9.9FVAHH398 pKa = 6.61LTPDD402 pKa = 3.2EE403 pKa = 4.42DD404 pKa = 4.09KK405 pKa = 11.42FMGGALKK412 pKa = 10.25ILNTAVPGPSGTSRR426 pKa = 11.84PPLDD430 pKa = 3.91IEE432 pKa = 4.72SEE434 pKa = 4.19
MM1 pKa = 7.57EE2 pKa = 6.3LYY4 pKa = 9.21NTIINGAKK12 pKa = 9.89EE13 pKa = 3.61IDD15 pKa = 3.45IGGSNPMTGALSGWTDD31 pKa = 3.38DD32 pKa = 5.05SYY34 pKa = 11.73KK35 pKa = 10.41QLYY38 pKa = 7.86GHH40 pKa = 6.46VRR42 pKa = 11.84ITQLEE47 pKa = 4.31VSFIPRR53 pKa = 11.84ILKK56 pKa = 9.9AWFSTYY62 pKa = 10.54YY63 pKa = 9.65NHH65 pKa = 7.61NIGNIDD71 pKa = 3.97LSSLMMSTLSLITVSSVSKK90 pKa = 10.38GQTSQTPNIQEE101 pKa = 4.17MISIAEE107 pKa = 3.94NLGKK111 pKa = 10.3VEE113 pKa = 3.99SQLRR117 pKa = 11.84KK118 pKa = 9.53NSPLIDD124 pKa = 4.01HH125 pKa = 7.39DD126 pKa = 4.53SEE128 pKa = 5.86KK129 pKa = 11.13EE130 pKa = 3.85DD131 pKa = 3.68ILAGDD136 pKa = 4.26HH137 pKa = 6.58EE138 pKa = 4.84EE139 pKa = 3.93QHH141 pKa = 6.96SIDD144 pKa = 4.17SSDD147 pKa = 3.36VMYY150 pKa = 11.07LSMISDD156 pKa = 4.36PDD158 pKa = 3.79YY159 pKa = 11.24QDD161 pKa = 3.12TLINIASFQALVMLRR176 pKa = 11.84HH177 pKa = 5.16ITKK180 pKa = 9.92TNTSVDD186 pKa = 3.06KK187 pKa = 10.58YY188 pKa = 10.55QRR190 pKa = 11.84EE191 pKa = 4.31SLTRR195 pKa = 11.84TLKK198 pKa = 10.64SLCPNTLLTADD209 pKa = 4.63IPPPSAQFVTYY220 pKa = 9.95FKK222 pKa = 10.6LNCMKK227 pKa = 10.7GNDD230 pKa = 3.55VATSLLAYY238 pKa = 10.38VMNLYY243 pKa = 9.53MEE245 pKa = 5.13ARR247 pKa = 11.84QSGATPLVDD256 pKa = 3.86YY257 pKa = 11.24LNAGCLTHH265 pKa = 7.25LKK267 pKa = 10.86GNGLALLEE275 pKa = 5.51LIFQVHH281 pKa = 6.89LCTKK285 pKa = 10.44LKK287 pKa = 10.87LSDD290 pKa = 4.47LFDD293 pKa = 3.47MTYY296 pKa = 10.88SSQCAQSIDD305 pKa = 3.29RR306 pKa = 11.84AYY308 pKa = 11.02HH309 pKa = 5.48HH310 pKa = 6.52FKK312 pKa = 10.57EE313 pKa = 4.22YY314 pKa = 11.4AEE316 pKa = 4.6LKK318 pKa = 10.84SNDD321 pKa = 3.17GKK323 pKa = 10.64IIRR326 pKa = 11.84PAQVTWWWARR336 pKa = 11.84LFNDD340 pKa = 4.37KK341 pKa = 11.04YY342 pKa = 11.15FMDD345 pKa = 3.58MSIRR349 pKa = 11.84NNYY352 pKa = 8.21QYY354 pKa = 11.04CLRR357 pKa = 11.84LACILTCYY365 pKa = 8.33QQQGSPLEE373 pKa = 4.01AAGFGTVSAKK383 pKa = 10.16EE384 pKa = 3.93LSKK387 pKa = 11.42HH388 pKa = 5.91NDD390 pKa = 2.9FATKK394 pKa = 9.9FVAHH398 pKa = 6.61LTPDD402 pKa = 3.2EE403 pKa = 4.42DD404 pKa = 4.09KK405 pKa = 11.42FMGGALKK412 pKa = 10.25ILNTAVPGPSGTSRR426 pKa = 11.84PPLDD430 pKa = 3.91IEE432 pKa = 4.72SEE434 pKa = 4.19
Molecular weight: 48.6 kDa
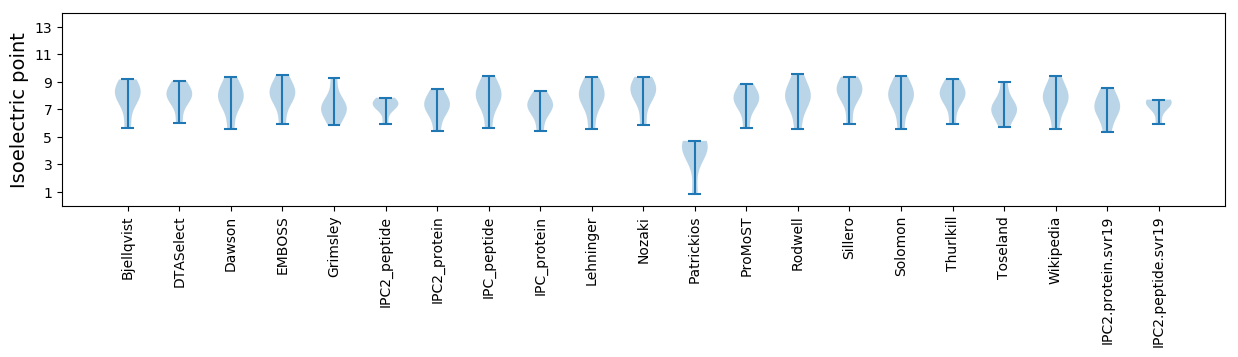
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0X9JET7|A0A0X9JET7_9RHAB Uncharacterized protein OS=Diachasmimorpha longicaudata rhabdovirus OX=1585246 GN=RS08_02 PE=4 SV=1
MM1 pKa = 6.88QQDD4 pKa = 2.97KK5 pKa = 10.13MGYY8 pKa = 9.07SVSFTLKK15 pKa = 10.15GHH17 pKa = 6.85LKK19 pKa = 10.28IINCPKK25 pKa = 9.66KK26 pKa = 9.45WLDD29 pKa = 3.47FKK31 pKa = 11.85YY32 pKa = 10.49EE33 pKa = 4.17LSCLILSHH41 pKa = 6.77WEE43 pKa = 3.7NDD45 pKa = 4.01FPCLLINEE53 pKa = 5.18EE54 pKa = 4.31IRR56 pKa = 11.84GLYY59 pKa = 10.29YY60 pKa = 10.73NVLVNLLEE68 pKa = 4.69KK69 pKa = 9.79KK70 pKa = 10.0HH71 pKa = 6.33VMIRR75 pKa = 11.84DD76 pKa = 3.54GVGGYY81 pKa = 10.04DD82 pKa = 4.19CYY84 pKa = 11.11IDD86 pKa = 5.54LISKK90 pKa = 9.96GFLHH94 pKa = 6.99KK95 pKa = 10.78AMRR98 pKa = 11.84QPVMTSVKK106 pKa = 9.49QEE108 pKa = 3.73SRR110 pKa = 11.84ITSVSFPLTEE120 pKa = 4.1DD121 pKa = 3.04TTILVKK127 pKa = 10.78GIIQLIQCPFKK138 pKa = 10.48IDD140 pKa = 3.16NWEE143 pKa = 3.85KK144 pKa = 10.92AVAQGYY150 pKa = 9.54RR151 pKa = 11.84PFNNMIKK158 pKa = 9.96KK159 pKa = 10.57DD160 pKa = 3.65SGGKK164 pKa = 9.41KK165 pKa = 9.49NQVEE169 pKa = 4.19LRR171 pKa = 11.84SISSGPSTAVPEE183 pKa = 4.32EE184 pKa = 3.96IKK186 pKa = 10.65IKK188 pKa = 10.53NLLYY192 pKa = 10.83NWIKK196 pKa = 10.71KK197 pKa = 8.84HH198 pKa = 5.6
MM1 pKa = 6.88QQDD4 pKa = 2.97KK5 pKa = 10.13MGYY8 pKa = 9.07SVSFTLKK15 pKa = 10.15GHH17 pKa = 6.85LKK19 pKa = 10.28IINCPKK25 pKa = 9.66KK26 pKa = 9.45WLDD29 pKa = 3.47FKK31 pKa = 11.85YY32 pKa = 10.49EE33 pKa = 4.17LSCLILSHH41 pKa = 6.77WEE43 pKa = 3.7NDD45 pKa = 4.01FPCLLINEE53 pKa = 5.18EE54 pKa = 4.31IRR56 pKa = 11.84GLYY59 pKa = 10.29YY60 pKa = 10.73NVLVNLLEE68 pKa = 4.69KK69 pKa = 9.79KK70 pKa = 10.0HH71 pKa = 6.33VMIRR75 pKa = 11.84DD76 pKa = 3.54GVGGYY81 pKa = 10.04DD82 pKa = 4.19CYY84 pKa = 11.11IDD86 pKa = 5.54LISKK90 pKa = 9.96GFLHH94 pKa = 6.99KK95 pKa = 10.78AMRR98 pKa = 11.84QPVMTSVKK106 pKa = 9.49QEE108 pKa = 3.73SRR110 pKa = 11.84ITSVSFPLTEE120 pKa = 4.1DD121 pKa = 3.04TTILVKK127 pKa = 10.78GIIQLIQCPFKK138 pKa = 10.48IDD140 pKa = 3.16NWEE143 pKa = 3.85KK144 pKa = 10.92AVAQGYY150 pKa = 9.54RR151 pKa = 11.84PFNNMIKK158 pKa = 9.96KK159 pKa = 10.57DD160 pKa = 3.65SGGKK164 pKa = 9.41KK165 pKa = 9.49NQVEE169 pKa = 4.19LRR171 pKa = 11.84SISSGPSTAVPEE183 pKa = 4.32EE184 pKa = 3.96IKK186 pKa = 10.65IKK188 pKa = 10.53NLLYY192 pKa = 10.83NWIKK196 pKa = 10.71KK197 pKa = 8.84HH198 pKa = 5.6
Molecular weight: 22.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3971 |
198 |
2110 |
661.8 |
75.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.651 ± 0.6 | 1.939 ± 0.089 |
4.684 ± 0.357 | 5.691 ± 0.645 |
4.18 ± 0.231 | 5.162 ± 0.139 |
2.871 ± 0.256 | 7.63 ± 0.21 |
6.573 ± 0.728 | 10.677 ± 1.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.846 ± 0.226 | 4.81 ± 0.217 |
4.911 ± 0.216 | 3.803 ± 0.221 |
4.13 ± 0.273 | 9.62 ± 0.47 |
5.993 ± 0.64 | 5.288 ± 0.55 |
1.637 ± 0.278 | 3.903 ± 0.257 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
