
Mycoplana dimorpha
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Brucellaceae; Mycoplana
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
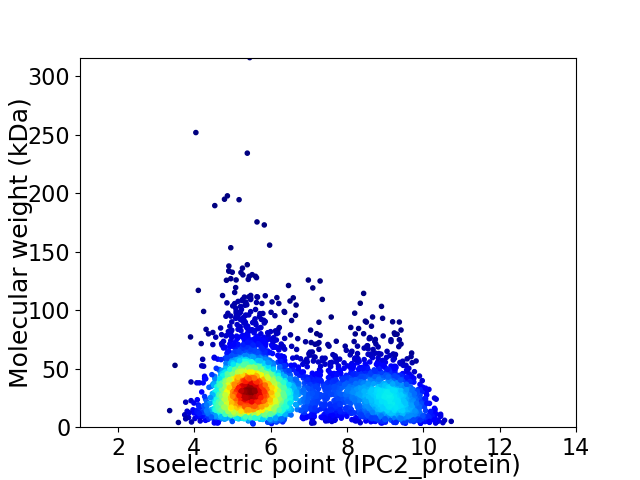
Virtual 2D-PAGE plot for 4217 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
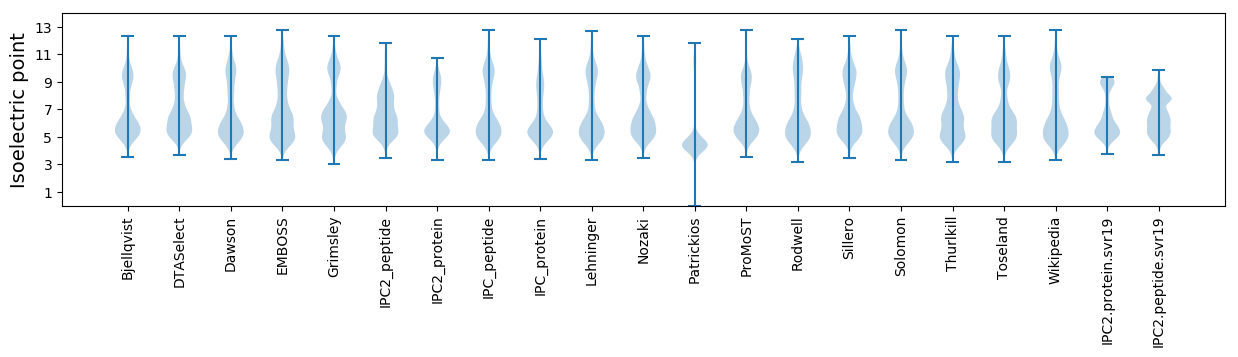
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T5BB28|A0A2T5BB28_MYCDI 2-oxoisovalerate dehydrogenase subunit alpha OS=Mycoplana dimorpha OX=28320 GN=C7449_103191 PE=3 SV=1
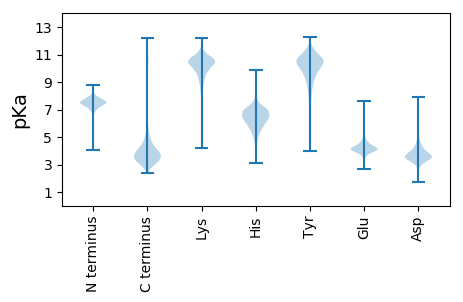
MM1 pKa = 7.48SMKK4 pKa = 10.41FLTSTALLALVAAPALAADD23 pKa = 3.74VVSYY27 pKa = 9.41QEE29 pKa = 4.11PAPQVEE35 pKa = 4.35ASPAFSWTGGYY46 pKa = 10.0VGLQGGGAWLNGDD59 pKa = 3.97FSIPGASASEE69 pKa = 4.17DD70 pKa = 3.69FNGGLIGGFAGYY82 pKa = 10.53NFQQGNWVFGVEE94 pKa = 4.59GDD96 pKa = 4.75VSYY99 pKa = 11.55NWNDD103 pKa = 2.96NNYY106 pKa = 10.61DD107 pKa = 3.32IFGTTAEE114 pKa = 4.28VGTDD118 pKa = 3.03VSGSVRR124 pKa = 11.84GRR126 pKa = 11.84AGYY129 pKa = 10.84AFDD132 pKa = 4.03SALIYY137 pKa = 9.43ATAGWTATRR146 pKa = 11.84GFVDD150 pKa = 3.65VPGFDD155 pKa = 4.23KK156 pKa = 10.38EE157 pKa = 4.7TEE159 pKa = 4.26VFNGWTIGAGADD171 pKa = 3.31YY172 pKa = 11.21AFTNNIFGRR181 pKa = 11.84AEE183 pKa = 3.47YY184 pKa = 9.99RR185 pKa = 11.84YY186 pKa = 10.58NDD188 pKa = 4.14FGDD191 pKa = 3.8KK192 pKa = 10.41DD193 pKa = 3.62IQGVNVDD200 pKa = 4.41LDD202 pKa = 3.57QHH204 pKa = 5.84QVTLGIGVKK213 pKa = 9.91FF214 pKa = 3.75
MM1 pKa = 7.48SMKK4 pKa = 10.41FLTSTALLALVAAPALAADD23 pKa = 3.74VVSYY27 pKa = 9.41QEE29 pKa = 4.11PAPQVEE35 pKa = 4.35ASPAFSWTGGYY46 pKa = 10.0VGLQGGGAWLNGDD59 pKa = 3.97FSIPGASASEE69 pKa = 4.17DD70 pKa = 3.69FNGGLIGGFAGYY82 pKa = 10.53NFQQGNWVFGVEE94 pKa = 4.59GDD96 pKa = 4.75VSYY99 pKa = 11.55NWNDD103 pKa = 2.96NNYY106 pKa = 10.61DD107 pKa = 3.32IFGTTAEE114 pKa = 4.28VGTDD118 pKa = 3.03VSGSVRR124 pKa = 11.84GRR126 pKa = 11.84AGYY129 pKa = 10.84AFDD132 pKa = 4.03SALIYY137 pKa = 9.43ATAGWTATRR146 pKa = 11.84GFVDD150 pKa = 3.65VPGFDD155 pKa = 4.23KK156 pKa = 10.38EE157 pKa = 4.7TEE159 pKa = 4.26VFNGWTIGAGADD171 pKa = 3.31YY172 pKa = 11.21AFTNNIFGRR181 pKa = 11.84AEE183 pKa = 3.47YY184 pKa = 9.99RR185 pKa = 11.84YY186 pKa = 10.58NDD188 pKa = 4.14FGDD191 pKa = 3.8KK192 pKa = 10.41DD193 pKa = 3.62IQGVNVDD200 pKa = 4.41LDD202 pKa = 3.57QHH204 pKa = 5.84QVTLGIGVKK213 pKa = 9.91FF214 pKa = 3.75
Molecular weight: 22.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T5AXF0|A0A2T5AXF0_MYCDI Molybdenum import ATP-binding protein ModC OS=Mycoplana dimorpha OX=28320 GN=modC PE=3 SV=1
MM1 pKa = 7.8RR2 pKa = 11.84PFALSKK8 pKa = 10.53AIRR11 pKa = 11.84PLALAATASLVAACQSSAPAPQASTAALPTMEE43 pKa = 5.7RR44 pKa = 11.84IALGANACWFKK55 pKa = 11.33SGDD58 pKa = 3.66AAFQAYY64 pKa = 9.5RR65 pKa = 11.84LAPEE69 pKa = 4.69LNSFSGKK76 pKa = 9.23PRR78 pKa = 11.84ILVVPRR84 pKa = 11.84RR85 pKa = 11.84SPEE88 pKa = 3.69ARR90 pKa = 11.84PLLVVQAEE98 pKa = 4.12GHH100 pKa = 5.96PARR103 pKa = 11.84LSAFGPLMDD112 pKa = 4.63EE113 pKa = 4.29PVSGRR118 pKa = 11.84IAADD122 pKa = 3.38VKK124 pKa = 10.16RR125 pKa = 11.84WAAGGAGCRR134 pKa = 3.99
MM1 pKa = 7.8RR2 pKa = 11.84PFALSKK8 pKa = 10.53AIRR11 pKa = 11.84PLALAATASLVAACQSSAPAPQASTAALPTMEE43 pKa = 5.7RR44 pKa = 11.84IALGANACWFKK55 pKa = 11.33SGDD58 pKa = 3.66AAFQAYY64 pKa = 9.5RR65 pKa = 11.84LAPEE69 pKa = 4.69LNSFSGKK76 pKa = 9.23PRR78 pKa = 11.84ILVVPRR84 pKa = 11.84RR85 pKa = 11.84SPEE88 pKa = 3.69ARR90 pKa = 11.84PLLVVQAEE98 pKa = 4.12GHH100 pKa = 5.96PARR103 pKa = 11.84LSAFGPLMDD112 pKa = 4.63EE113 pKa = 4.29PVSGRR118 pKa = 11.84IAADD122 pKa = 3.38VKK124 pKa = 10.16RR125 pKa = 11.84WAAGGAGCRR134 pKa = 3.99
Molecular weight: 13.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1319512 |
26 |
2827 |
312.9 |
33.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.453 ± 0.05 | 0.832 ± 0.014 |
5.633 ± 0.029 | 5.832 ± 0.034 |
3.843 ± 0.02 | 8.524 ± 0.037 |
2.032 ± 0.019 | 5.379 ± 0.027 |
3.4 ± 0.031 | 10.004 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.552 ± 0.017 | 2.662 ± 0.022 |
4.999 ± 0.027 | 3.024 ± 0.023 |
7.07 ± 0.041 | 5.425 ± 0.023 |
5.236 ± 0.028 | 7.514 ± 0.03 |
1.28 ± 0.016 | 2.306 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
