
Rattus norvegicus polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
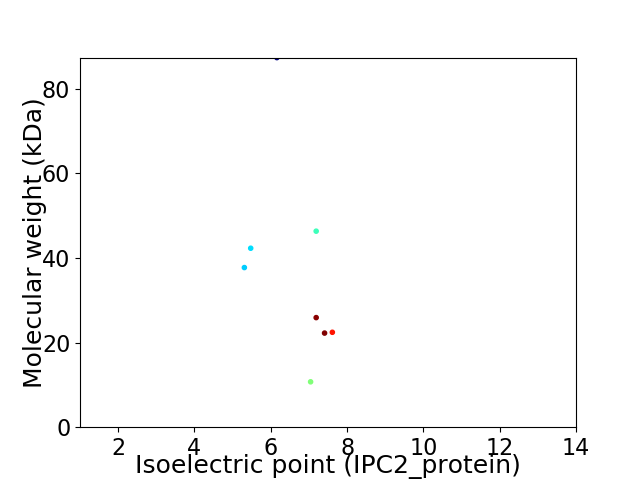
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4PP86|A0A0H4PP86_9POLY Capsid protein VP1 OS=Rattus norvegicus polyomavirus 1 OX=1679933 GN=VP1 PE=3 SV=1
MM1 pKa = 7.29GAAISVIIDD10 pKa = 4.47LIATATEE17 pKa = 4.56LSTATGFTVEE27 pKa = 4.41AVLSGEE33 pKa = 3.88ALAAIEE39 pKa = 4.38AQVSTIATMEE49 pKa = 4.06GLAAEE54 pKa = 4.41SALASMGITPEE65 pKa = 4.0MYY67 pKa = 11.15AMISTVPPTVEE78 pKa = 4.68GIISGAAGTAHH89 pKa = 7.69ILQTIAGSASLTLGVISGFRR109 pKa = 11.84DD110 pKa = 3.45QQVSIVNRR118 pKa = 11.84NMALIPWRR126 pKa = 11.84PEE128 pKa = 3.79EE129 pKa = 4.05YY130 pKa = 9.79WDD132 pKa = 3.86VLFPGVNTIARR143 pKa = 11.84GLDD146 pKa = 3.61VAHH149 pKa = 6.87GWAHH153 pKa = 6.69SLFQSVSRR161 pKa = 11.84QIWRR165 pKa = 11.84TLFEE169 pKa = 4.15EE170 pKa = 4.03TRR172 pKa = 11.84AAIDD176 pKa = 3.45YY177 pKa = 9.81AVRR180 pKa = 11.84DD181 pKa = 3.55VTIRR185 pKa = 11.84QTHH188 pKa = 6.13RR189 pKa = 11.84LLDD192 pKa = 3.69GVARR196 pKa = 11.84MLEE199 pKa = 3.9NTRR202 pKa = 11.84WVVTNAVEE210 pKa = 3.87QGGQIVSDD218 pKa = 3.92AVQNTGQLVLTAPGEE233 pKa = 4.24AYY235 pKa = 10.18RR236 pKa = 11.84GLRR239 pKa = 11.84NYY241 pKa = 9.9YY242 pKa = 10.73AEE244 pKa = 4.5LPGVNPAQRR253 pKa = 11.84QQMLRR258 pKa = 11.84TIEE261 pKa = 4.16RR262 pKa = 11.84EE263 pKa = 3.76NAQQALRR270 pKa = 11.84TEE272 pKa = 4.3VGRR275 pKa = 11.84DD276 pKa = 3.61VQSVTPSPKK285 pKa = 9.85SSGFTEE291 pKa = 4.13SGNVIQRR298 pKa = 11.84YY299 pKa = 5.76MPPGGAMQRR308 pKa = 11.84TCPDD312 pKa = 2.37WMLPLILGLYY322 pKa = 10.36GDD324 pKa = 5.34INPTWHH330 pKa = 6.46SVIEE334 pKa = 4.21EE335 pKa = 4.29EE336 pKa = 5.24YY337 pKa = 10.82GPQKK341 pKa = 10.17KK342 pKa = 9.2RR343 pKa = 11.84RR344 pKa = 11.84RR345 pKa = 11.84LQQ347 pKa = 3.2
MM1 pKa = 7.29GAAISVIIDD10 pKa = 4.47LIATATEE17 pKa = 4.56LSTATGFTVEE27 pKa = 4.41AVLSGEE33 pKa = 3.88ALAAIEE39 pKa = 4.38AQVSTIATMEE49 pKa = 4.06GLAAEE54 pKa = 4.41SALASMGITPEE65 pKa = 4.0MYY67 pKa = 11.15AMISTVPPTVEE78 pKa = 4.68GIISGAAGTAHH89 pKa = 7.69ILQTIAGSASLTLGVISGFRR109 pKa = 11.84DD110 pKa = 3.45QQVSIVNRR118 pKa = 11.84NMALIPWRR126 pKa = 11.84PEE128 pKa = 3.79EE129 pKa = 4.05YY130 pKa = 9.79WDD132 pKa = 3.86VLFPGVNTIARR143 pKa = 11.84GLDD146 pKa = 3.61VAHH149 pKa = 6.87GWAHH153 pKa = 6.69SLFQSVSRR161 pKa = 11.84QIWRR165 pKa = 11.84TLFEE169 pKa = 4.15EE170 pKa = 4.03TRR172 pKa = 11.84AAIDD176 pKa = 3.45YY177 pKa = 9.81AVRR180 pKa = 11.84DD181 pKa = 3.55VTIRR185 pKa = 11.84QTHH188 pKa = 6.13RR189 pKa = 11.84LLDD192 pKa = 3.69GVARR196 pKa = 11.84MLEE199 pKa = 3.9NTRR202 pKa = 11.84WVVTNAVEE210 pKa = 3.87QGGQIVSDD218 pKa = 3.92AVQNTGQLVLTAPGEE233 pKa = 4.24AYY235 pKa = 10.18RR236 pKa = 11.84GLRR239 pKa = 11.84NYY241 pKa = 9.9YY242 pKa = 10.73AEE244 pKa = 4.5LPGVNPAQRR253 pKa = 11.84QQMLRR258 pKa = 11.84TIEE261 pKa = 4.16RR262 pKa = 11.84EE263 pKa = 3.76NAQQALRR270 pKa = 11.84TEE272 pKa = 4.3VGRR275 pKa = 11.84DD276 pKa = 3.61VQSVTPSPKK285 pKa = 9.85SSGFTEE291 pKa = 4.13SGNVIQRR298 pKa = 11.84YY299 pKa = 5.76MPPGGAMQRR308 pKa = 11.84TCPDD312 pKa = 2.37WMLPLILGLYY322 pKa = 10.36GDD324 pKa = 5.34INPTWHH330 pKa = 6.46SVIEE334 pKa = 4.21EE335 pKa = 4.29EE336 pKa = 5.24YY337 pKa = 10.82GPQKK341 pKa = 10.17KK342 pKa = 9.2RR343 pKa = 11.84RR344 pKa = 11.84RR345 pKa = 11.84LQQ347 pKa = 3.2
Molecular weight: 37.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4PK86|A0A0H4PK86_9POLY Minor capsid protein OS=Rattus norvegicus polyomavirus 1 OX=1679933 GN=VP3 PE=3 SV=1
MM1 pKa = 7.92DD2 pKa = 5.99KK3 pKa = 10.69ILTKK7 pKa = 9.86EE8 pKa = 4.2EE9 pKa = 4.1KK10 pKa = 10.27SQLLALLDD18 pKa = 4.57LDD20 pKa = 3.79GQYY23 pKa = 10.18WGDD26 pKa = 3.82FGRR29 pKa = 11.84MQKK32 pKa = 10.48AYY34 pKa = 9.76HH35 pKa = 5.52AQSLKK40 pKa = 10.25FHH42 pKa = 7.28PDD44 pKa = 2.83KK45 pKa = 11.59GGDD48 pKa = 3.7PLLMQLLNTLWTKK61 pKa = 10.61LKK63 pKa = 10.64HH64 pKa = 5.97GLHH67 pKa = 5.02QVRR70 pKa = 11.84LNLGGHH76 pKa = 5.26EE77 pKa = 4.21VRR79 pKa = 11.84RR80 pKa = 11.84LPEE83 pKa = 4.3GDD85 pKa = 2.47WWLSTRR91 pKa = 11.84QTFGGNYY98 pKa = 8.69SRR100 pKa = 11.84RR101 pKa = 11.84LCRR104 pKa = 11.84MPITCLQNKK113 pKa = 9.21ACSTCNCILCTLRR126 pKa = 11.84RR127 pKa = 11.84QHH129 pKa = 6.94RR130 pKa = 11.84GLKK133 pKa = 9.2KK134 pKa = 10.57ACLAPCLVLGEE145 pKa = 5.05CYY147 pKa = 10.6CLDD150 pKa = 4.78CFSLWFGLPINNFIVVLYY168 pKa = 11.02AEE170 pKa = 4.74FLGDD174 pKa = 5.62IPVDD178 pKa = 3.13WLDD181 pKa = 3.39IDD183 pKa = 3.55VHH185 pKa = 6.81QIYY188 pKa = 10.56NPAVGKK194 pKa = 9.97YY195 pKa = 8.83
MM1 pKa = 7.92DD2 pKa = 5.99KK3 pKa = 10.69ILTKK7 pKa = 9.86EE8 pKa = 4.2EE9 pKa = 4.1KK10 pKa = 10.27SQLLALLDD18 pKa = 4.57LDD20 pKa = 3.79GQYY23 pKa = 10.18WGDD26 pKa = 3.82FGRR29 pKa = 11.84MQKK32 pKa = 10.48AYY34 pKa = 9.76HH35 pKa = 5.52AQSLKK40 pKa = 10.25FHH42 pKa = 7.28PDD44 pKa = 2.83KK45 pKa = 11.59GGDD48 pKa = 3.7PLLMQLLNTLWTKK61 pKa = 10.61LKK63 pKa = 10.64HH64 pKa = 5.97GLHH67 pKa = 5.02QVRR70 pKa = 11.84LNLGGHH76 pKa = 5.26EE77 pKa = 4.21VRR79 pKa = 11.84RR80 pKa = 11.84LPEE83 pKa = 4.3GDD85 pKa = 2.47WWLSTRR91 pKa = 11.84QTFGGNYY98 pKa = 8.69SRR100 pKa = 11.84RR101 pKa = 11.84LCRR104 pKa = 11.84MPITCLQNKK113 pKa = 9.21ACSTCNCILCTLRR126 pKa = 11.84RR127 pKa = 11.84QHH129 pKa = 6.94RR130 pKa = 11.84GLKK133 pKa = 9.2KK134 pKa = 10.57ACLAPCLVLGEE145 pKa = 5.05CYY147 pKa = 10.6CLDD150 pKa = 4.78CFSLWFGLPINNFIVVLYY168 pKa = 11.02AEE170 pKa = 4.74FLGDD174 pKa = 5.62IPVDD178 pKa = 3.13WLDD181 pKa = 3.39IDD183 pKa = 3.55VHH185 pKa = 6.81QIYY188 pKa = 10.56NPAVGKK194 pKa = 9.97YY195 pKa = 8.83
Molecular weight: 22.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2625 |
95 |
776 |
328.1 |
36.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.829 ± 0.797 | 2.629 ± 0.666 |
5.143 ± 0.446 | 5.371 ± 0.387 |
3.2 ± 0.714 | 7.619 ± 0.671 |
2.667 ± 0.446 | 4.724 ± 0.578 |
4.914 ± 0.992 | 11.086 ± 1.16 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.514 ± 0.28 | 4.571 ± 0.517 |
6.286 ± 0.522 | 5.41 ± 0.302 |
5.524 ± 0.673 | 6.019 ± 0.484 |
5.562 ± 0.508 | 5.6 ± 0.668 |
2.057 ± 0.226 | 3.276 ± 0.321 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
