
Hubei mosquito virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.81
Get precalculated fractions of proteins
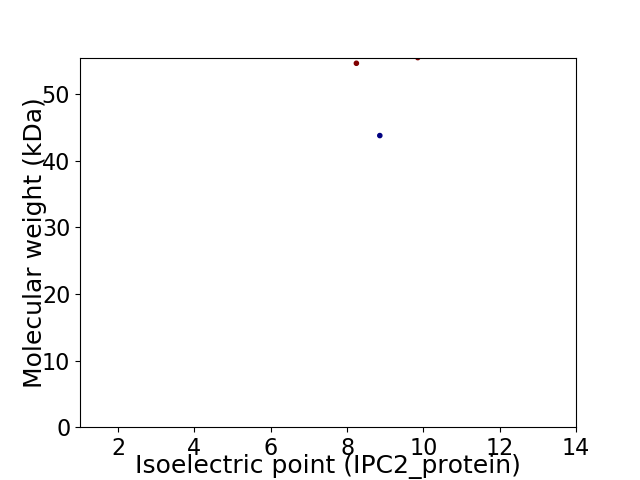
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFE1|A0A1L3KFE1_9VIRU Uncharacterized protein OS=Hubei mosquito virus 4 OX=1922928 PE=4 SV=1
MM1 pKa = 7.79KK2 pKa = 10.13GRR4 pKa = 11.84AISSSQLPGSSLRR17 pKa = 11.84TGEE20 pKa = 4.77CGHH23 pKa = 5.42SCKK26 pKa = 10.43RR27 pKa = 11.84YY28 pKa = 6.36TRR30 pKa = 11.84KK31 pKa = 9.2MFDD34 pKa = 3.12YY35 pKa = 9.18DD36 pKa = 3.39TDD38 pKa = 4.03FGGSGSVVKK47 pKa = 8.78THH49 pKa = 6.91LSCVCNEE56 pKa = 3.83VLALMNRR63 pKa = 11.84HH64 pKa = 5.58QLDD67 pKa = 3.43DD68 pKa = 3.69GARR71 pKa = 11.84YY72 pKa = 7.66TFTGDD77 pKa = 2.76LTKK80 pKa = 9.92WVRR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84VRR87 pKa = 11.84PMYY90 pKa = 9.36PWSEE94 pKa = 4.3SMVIEE99 pKa = 4.47HH100 pKa = 7.78AIPSKK105 pKa = 10.76KK106 pKa = 9.96KK107 pKa = 10.21LLISAKK113 pKa = 9.76EE114 pKa = 4.06SLSKK118 pKa = 11.28YY119 pKa = 9.88PLTASDD125 pKa = 4.12GAIKK129 pKa = 10.18MFLKK133 pKa = 10.34DD134 pKa = 3.72DD135 pKa = 4.18KK136 pKa = 11.71YY137 pKa = 10.79HH138 pKa = 7.37GEE140 pKa = 4.22LKK142 pKa = 10.53DD143 pKa = 3.65PRR145 pKa = 11.84CIQYY149 pKa = 10.24RR150 pKa = 11.84NKK152 pKa = 10.62RR153 pKa = 11.84YY154 pKa = 9.68GLRR157 pKa = 11.84LATYY161 pKa = 7.44LHH163 pKa = 6.68PLEE166 pKa = 5.23KK167 pKa = 10.73YY168 pKa = 9.5VMSWTRR174 pKa = 11.84NGTHH178 pKa = 6.79IFAKK182 pKa = 9.08GRR184 pKa = 11.84NMRR187 pKa = 11.84QRR189 pKa = 11.84GRR191 pKa = 11.84DD192 pKa = 3.06IAHH195 pKa = 6.92KK196 pKa = 9.78FNRR199 pKa = 11.84YY200 pKa = 9.03DD201 pKa = 3.15EE202 pKa = 4.62CAVVSLDD209 pKa = 3.44HH210 pKa = 7.25SKK212 pKa = 10.71FDD214 pKa = 3.54CHH216 pKa = 7.29VNEE219 pKa = 4.09SLLRR223 pKa = 11.84AEE225 pKa = 3.93HH226 pKa = 6.41WFYY229 pKa = 10.27NQCYY233 pKa = 9.52GSEE236 pKa = 3.98EE237 pKa = 4.51LEE239 pKa = 4.32FLLEE243 pKa = 3.89LQVRR247 pKa = 11.84NKK249 pKa = 10.53GMTKK253 pKa = 10.41NGTKK257 pKa = 8.92YY258 pKa = 5.78TTRR261 pKa = 11.84ATRR264 pKa = 11.84MSGDD268 pKa = 3.33QNTGLGNCLINYY280 pKa = 7.48MMTKK284 pKa = 10.0QLLSKK289 pKa = 10.84LGIGYY294 pKa = 9.68DD295 pKa = 3.83LYY297 pKa = 10.84IDD299 pKa = 4.76GDD301 pKa = 3.9DD302 pKa = 4.12FLIFMEE308 pKa = 4.68KK309 pKa = 9.78RR310 pKa = 11.84HH311 pKa = 5.94VPLIDD316 pKa = 3.64PSAYY320 pKa = 10.33SVFGMSTKK328 pKa = 10.51LEE330 pKa = 4.27SVAYY334 pKa = 8.82TIEE337 pKa = 4.49HH338 pKa = 6.67VDD340 pKa = 4.23FCQCRR345 pKa = 11.84PVFDD349 pKa = 3.97GVGYY353 pKa = 9.2TMVRR357 pKa = 11.84NPEE360 pKa = 3.78RR361 pKa = 11.84MLARR365 pKa = 11.84LPWIVGPVEE374 pKa = 4.04DD375 pKa = 4.98SRR377 pKa = 11.84ANDD380 pKa = 3.13IVFSTGQCEE389 pKa = 3.96IAQGLGLPIGQYY401 pKa = 9.48IGQRR405 pKa = 11.84MCEE408 pKa = 3.79RR409 pKa = 11.84GGKK412 pKa = 6.99MVRR415 pKa = 11.84IRR417 pKa = 11.84HH418 pKa = 5.37RR419 pKa = 11.84AWLEE423 pKa = 3.52KK424 pKa = 9.79MKK426 pKa = 10.25PGKK429 pKa = 9.45LQPIEE434 pKa = 3.79PMGGVRR440 pKa = 11.84EE441 pKa = 4.26SYY443 pKa = 9.9EE444 pKa = 4.21RR445 pKa = 11.84AWGLSIADD453 pKa = 3.66QLLIEE458 pKa = 4.3QTSLVEE464 pKa = 4.2PEE466 pKa = 3.81EE467 pKa = 4.23HH468 pKa = 6.38FAPGILQWW476 pKa = 3.74
MM1 pKa = 7.79KK2 pKa = 10.13GRR4 pKa = 11.84AISSSQLPGSSLRR17 pKa = 11.84TGEE20 pKa = 4.77CGHH23 pKa = 5.42SCKK26 pKa = 10.43RR27 pKa = 11.84YY28 pKa = 6.36TRR30 pKa = 11.84KK31 pKa = 9.2MFDD34 pKa = 3.12YY35 pKa = 9.18DD36 pKa = 3.39TDD38 pKa = 4.03FGGSGSVVKK47 pKa = 8.78THH49 pKa = 6.91LSCVCNEE56 pKa = 3.83VLALMNRR63 pKa = 11.84HH64 pKa = 5.58QLDD67 pKa = 3.43DD68 pKa = 3.69GARR71 pKa = 11.84YY72 pKa = 7.66TFTGDD77 pKa = 2.76LTKK80 pKa = 9.92WVRR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84VRR87 pKa = 11.84PMYY90 pKa = 9.36PWSEE94 pKa = 4.3SMVIEE99 pKa = 4.47HH100 pKa = 7.78AIPSKK105 pKa = 10.76KK106 pKa = 9.96KK107 pKa = 10.21LLISAKK113 pKa = 9.76EE114 pKa = 4.06SLSKK118 pKa = 11.28YY119 pKa = 9.88PLTASDD125 pKa = 4.12GAIKK129 pKa = 10.18MFLKK133 pKa = 10.34DD134 pKa = 3.72DD135 pKa = 4.18KK136 pKa = 11.71YY137 pKa = 10.79HH138 pKa = 7.37GEE140 pKa = 4.22LKK142 pKa = 10.53DD143 pKa = 3.65PRR145 pKa = 11.84CIQYY149 pKa = 10.24RR150 pKa = 11.84NKK152 pKa = 10.62RR153 pKa = 11.84YY154 pKa = 9.68GLRR157 pKa = 11.84LATYY161 pKa = 7.44LHH163 pKa = 6.68PLEE166 pKa = 5.23KK167 pKa = 10.73YY168 pKa = 9.5VMSWTRR174 pKa = 11.84NGTHH178 pKa = 6.79IFAKK182 pKa = 9.08GRR184 pKa = 11.84NMRR187 pKa = 11.84QRR189 pKa = 11.84GRR191 pKa = 11.84DD192 pKa = 3.06IAHH195 pKa = 6.92KK196 pKa = 9.78FNRR199 pKa = 11.84YY200 pKa = 9.03DD201 pKa = 3.15EE202 pKa = 4.62CAVVSLDD209 pKa = 3.44HH210 pKa = 7.25SKK212 pKa = 10.71FDD214 pKa = 3.54CHH216 pKa = 7.29VNEE219 pKa = 4.09SLLRR223 pKa = 11.84AEE225 pKa = 3.93HH226 pKa = 6.41WFYY229 pKa = 10.27NQCYY233 pKa = 9.52GSEE236 pKa = 3.98EE237 pKa = 4.51LEE239 pKa = 4.32FLLEE243 pKa = 3.89LQVRR247 pKa = 11.84NKK249 pKa = 10.53GMTKK253 pKa = 10.41NGTKK257 pKa = 8.92YY258 pKa = 5.78TTRR261 pKa = 11.84ATRR264 pKa = 11.84MSGDD268 pKa = 3.33QNTGLGNCLINYY280 pKa = 7.48MMTKK284 pKa = 10.0QLLSKK289 pKa = 10.84LGIGYY294 pKa = 9.68DD295 pKa = 3.83LYY297 pKa = 10.84IDD299 pKa = 4.76GDD301 pKa = 3.9DD302 pKa = 4.12FLIFMEE308 pKa = 4.68KK309 pKa = 9.78RR310 pKa = 11.84HH311 pKa = 5.94VPLIDD316 pKa = 3.64PSAYY320 pKa = 10.33SVFGMSTKK328 pKa = 10.51LEE330 pKa = 4.27SVAYY334 pKa = 8.82TIEE337 pKa = 4.49HH338 pKa = 6.67VDD340 pKa = 4.23FCQCRR345 pKa = 11.84PVFDD349 pKa = 3.97GVGYY353 pKa = 9.2TMVRR357 pKa = 11.84NPEE360 pKa = 3.78RR361 pKa = 11.84MLARR365 pKa = 11.84LPWIVGPVEE374 pKa = 4.04DD375 pKa = 4.98SRR377 pKa = 11.84ANDD380 pKa = 3.13IVFSTGQCEE389 pKa = 3.96IAQGLGLPIGQYY401 pKa = 9.48IGQRR405 pKa = 11.84MCEE408 pKa = 3.79RR409 pKa = 11.84GGKK412 pKa = 6.99MVRR415 pKa = 11.84IRR417 pKa = 11.84HH418 pKa = 5.37RR419 pKa = 11.84AWLEE423 pKa = 3.52KK424 pKa = 9.79MKK426 pKa = 10.25PGKK429 pKa = 9.45LQPIEE434 pKa = 3.79PMGGVRR440 pKa = 11.84EE441 pKa = 4.26SYY443 pKa = 9.9EE444 pKa = 4.21RR445 pKa = 11.84AWGLSIADD453 pKa = 3.66QLLIEE458 pKa = 4.3QTSLVEE464 pKa = 4.2PEE466 pKa = 3.81EE467 pKa = 4.23HH468 pKa = 6.38FAPGILQWW476 pKa = 3.74
Molecular weight: 54.59 kDa
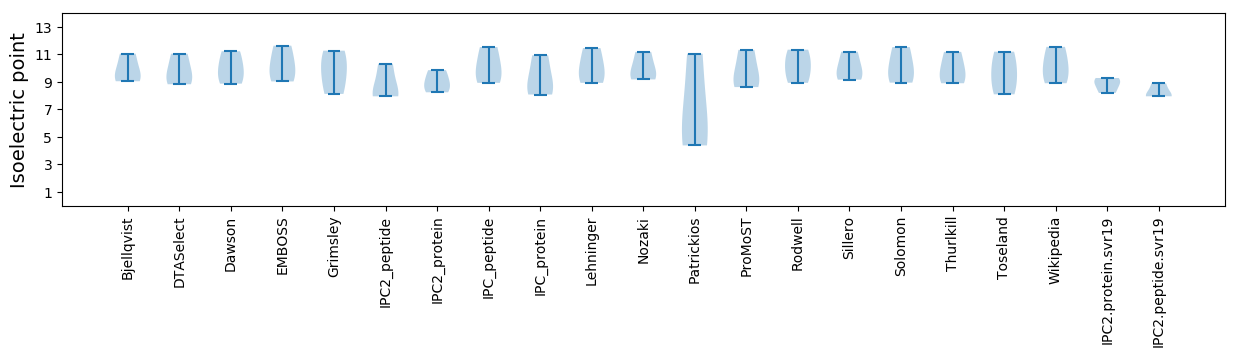
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFL5|A0A1L3KFL5_9VIRU Uncharacterized protein OS=Hubei mosquito virus 4 OX=1922928 PE=4 SV=1
MM1 pKa = 7.0SQHH4 pKa = 6.3GARR7 pKa = 11.84GQPRR11 pKa = 11.84GAGRR15 pKa = 11.84PTPTSGHH22 pKa = 6.2RR23 pKa = 11.84GKK25 pKa = 10.49DD26 pKa = 3.34GSKK29 pKa = 10.29EE30 pKa = 3.93PKK32 pKa = 10.31GDD34 pKa = 3.44DD35 pKa = 3.18RR36 pKa = 11.84QRR38 pKa = 11.84NPRR41 pKa = 11.84SAKK44 pKa = 10.06GKK46 pKa = 8.7PSGNARR52 pKa = 11.84GPQWQTVGRR61 pKa = 11.84KK62 pKa = 8.51PRR64 pKa = 11.84AANPEE69 pKa = 3.78ALLRR73 pKa = 11.84RR74 pKa = 11.84IHH76 pKa = 7.0ALEE79 pKa = 3.7QANRR83 pKa = 11.84NLLLQLGRR91 pKa = 11.84VNAGASGSGSTSRR104 pKa = 11.84GSTSGRR110 pKa = 11.84TNPASPARR118 pKa = 11.84PKK120 pKa = 9.87PAPRR124 pKa = 11.84TKK126 pKa = 10.7APAPAPRR133 pKa = 11.84TKK135 pKa = 10.77APAPPQRR142 pKa = 11.84PKK144 pKa = 11.07GPRR147 pKa = 11.84KK148 pKa = 9.31PLVRR152 pKa = 11.84GPGNRR157 pKa = 11.84TVSSSHH163 pKa = 6.13LPAVGTKK170 pKa = 10.47NSFQLLGEE178 pKa = 4.39EE179 pKa = 4.8FPHH182 pKa = 7.31LSRR185 pKa = 11.84GTLALAVSAGLSGVCTPTATIANGVQAEE213 pKa = 4.21ASTSGRR219 pKa = 11.84SPRR222 pKa = 11.84STRR225 pKa = 11.84RR226 pKa = 11.84ANSSPEE232 pKa = 3.9LNVRR236 pKa = 11.84RR237 pKa = 11.84PAPRR241 pKa = 11.84AEE243 pKa = 4.11GQRR246 pKa = 11.84RR247 pKa = 11.84SPASRR252 pKa = 11.84TNSIPTQGSSSQHH265 pKa = 5.44RR266 pKa = 11.84AATAEE271 pKa = 4.36PKK273 pKa = 9.99VAPNPAGSDD282 pKa = 3.16RR283 pKa = 11.84VPQPRR288 pKa = 11.84LRR290 pKa = 11.84MKK292 pKa = 10.55PGDD295 pKa = 4.32HH296 pKa = 6.19GWKK299 pKa = 10.09GAGPPGVFHH308 pKa = 7.06SPTVVSPRR316 pKa = 11.84SAGVCLEE323 pKa = 3.92VSAKK327 pKa = 10.32KK328 pKa = 10.9GEE330 pKa = 4.22LVEE333 pKa = 4.13NTNPVFVHH341 pKa = 6.57KK342 pKa = 10.19ATVRR346 pKa = 11.84KK347 pKa = 9.84DD348 pKa = 3.43VEE350 pKa = 4.64CGVGTTRR357 pKa = 11.84ANLAKK362 pKa = 10.75LLTTKK367 pKa = 9.73EE368 pKa = 3.79VSMAEE373 pKa = 4.59LYY375 pKa = 10.57RR376 pKa = 11.84DD377 pKa = 3.29ATVPMSSWKK386 pKa = 9.1DD387 pKa = 3.36TFGITIDD394 pKa = 3.34AAKK397 pKa = 10.54ALVVDD402 pKa = 4.03EE403 pKa = 4.22FLYY406 pKa = 11.17YY407 pKa = 10.52EE408 pKa = 4.19LVSRR412 pKa = 11.84FPFAVRR418 pKa = 11.84TADD421 pKa = 3.66LARR424 pKa = 11.84KK425 pKa = 7.97MFHH428 pKa = 6.87HH429 pKa = 7.23LNTVLAQYY437 pKa = 10.66DD438 pKa = 3.91CRR440 pKa = 11.84LYY442 pKa = 9.74TAKK445 pKa = 10.35EE446 pKa = 3.94LYY448 pKa = 9.76KK449 pKa = 10.69LKK451 pKa = 10.5QATVRR456 pKa = 11.84AALLPPAEE464 pKa = 4.24EE465 pKa = 4.01LLTRR469 pKa = 11.84KK470 pKa = 10.16LIQQEE475 pKa = 3.67ASQMEE480 pKa = 5.01KK481 pKa = 10.74YY482 pKa = 10.92NKK484 pKa = 9.95FADD487 pKa = 3.72KK488 pKa = 11.0GDD490 pKa = 3.94AGSVCRR496 pKa = 11.84PGSATASAFSSLLQSRR512 pKa = 11.84VGLRR516 pKa = 11.84PTPKK520 pKa = 10.41
MM1 pKa = 7.0SQHH4 pKa = 6.3GARR7 pKa = 11.84GQPRR11 pKa = 11.84GAGRR15 pKa = 11.84PTPTSGHH22 pKa = 6.2RR23 pKa = 11.84GKK25 pKa = 10.49DD26 pKa = 3.34GSKK29 pKa = 10.29EE30 pKa = 3.93PKK32 pKa = 10.31GDD34 pKa = 3.44DD35 pKa = 3.18RR36 pKa = 11.84QRR38 pKa = 11.84NPRR41 pKa = 11.84SAKK44 pKa = 10.06GKK46 pKa = 8.7PSGNARR52 pKa = 11.84GPQWQTVGRR61 pKa = 11.84KK62 pKa = 8.51PRR64 pKa = 11.84AANPEE69 pKa = 3.78ALLRR73 pKa = 11.84RR74 pKa = 11.84IHH76 pKa = 7.0ALEE79 pKa = 3.7QANRR83 pKa = 11.84NLLLQLGRR91 pKa = 11.84VNAGASGSGSTSRR104 pKa = 11.84GSTSGRR110 pKa = 11.84TNPASPARR118 pKa = 11.84PKK120 pKa = 9.87PAPRR124 pKa = 11.84TKK126 pKa = 10.7APAPAPRR133 pKa = 11.84TKK135 pKa = 10.77APAPPQRR142 pKa = 11.84PKK144 pKa = 11.07GPRR147 pKa = 11.84KK148 pKa = 9.31PLVRR152 pKa = 11.84GPGNRR157 pKa = 11.84TVSSSHH163 pKa = 6.13LPAVGTKK170 pKa = 10.47NSFQLLGEE178 pKa = 4.39EE179 pKa = 4.8FPHH182 pKa = 7.31LSRR185 pKa = 11.84GTLALAVSAGLSGVCTPTATIANGVQAEE213 pKa = 4.21ASTSGRR219 pKa = 11.84SPRR222 pKa = 11.84STRR225 pKa = 11.84RR226 pKa = 11.84ANSSPEE232 pKa = 3.9LNVRR236 pKa = 11.84RR237 pKa = 11.84PAPRR241 pKa = 11.84AEE243 pKa = 4.11GQRR246 pKa = 11.84RR247 pKa = 11.84SPASRR252 pKa = 11.84TNSIPTQGSSSQHH265 pKa = 5.44RR266 pKa = 11.84AATAEE271 pKa = 4.36PKK273 pKa = 9.99VAPNPAGSDD282 pKa = 3.16RR283 pKa = 11.84VPQPRR288 pKa = 11.84LRR290 pKa = 11.84MKK292 pKa = 10.55PGDD295 pKa = 4.32HH296 pKa = 6.19GWKK299 pKa = 10.09GAGPPGVFHH308 pKa = 7.06SPTVVSPRR316 pKa = 11.84SAGVCLEE323 pKa = 3.92VSAKK327 pKa = 10.32KK328 pKa = 10.9GEE330 pKa = 4.22LVEE333 pKa = 4.13NTNPVFVHH341 pKa = 6.57KK342 pKa = 10.19ATVRR346 pKa = 11.84KK347 pKa = 9.84DD348 pKa = 3.43VEE350 pKa = 4.64CGVGTTRR357 pKa = 11.84ANLAKK362 pKa = 10.75LLTTKK367 pKa = 9.73EE368 pKa = 3.79VSMAEE373 pKa = 4.59LYY375 pKa = 10.57RR376 pKa = 11.84DD377 pKa = 3.29ATVPMSSWKK386 pKa = 9.1DD387 pKa = 3.36TFGITIDD394 pKa = 3.34AAKK397 pKa = 10.54ALVVDD402 pKa = 4.03EE403 pKa = 4.22FLYY406 pKa = 11.17YY407 pKa = 10.52EE408 pKa = 4.19LVSRR412 pKa = 11.84FPFAVRR418 pKa = 11.84TADD421 pKa = 3.66LARR424 pKa = 11.84KK425 pKa = 7.97MFHH428 pKa = 6.87HH429 pKa = 7.23LNTVLAQYY437 pKa = 10.66DD438 pKa = 3.91CRR440 pKa = 11.84LYY442 pKa = 9.74TAKK445 pKa = 10.35EE446 pKa = 3.94LYY448 pKa = 9.76KK449 pKa = 10.69LKK451 pKa = 10.5QATVRR456 pKa = 11.84AALLPPAEE464 pKa = 4.24EE465 pKa = 4.01LLTRR469 pKa = 11.84KK470 pKa = 10.16LIQQEE475 pKa = 3.67ASQMEE480 pKa = 5.01KK481 pKa = 10.74YY482 pKa = 10.92NKK484 pKa = 9.95FADD487 pKa = 3.72KK488 pKa = 11.0GDD490 pKa = 3.94AGSVCRR496 pKa = 11.84PGSATASAFSSLLQSRR512 pKa = 11.84VGLRR516 pKa = 11.84PTPKK520 pKa = 10.41
Molecular weight: 55.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1396 |
400 |
520 |
465.3 |
51.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.166 ± 2.07 | 1.504 ± 0.549 |
4.155 ± 0.75 | 5.014 ± 0.528 |
3.51 ± 0.747 | 8.453 ± 0.098 |
2.077 ± 0.516 | 3.08 ± 1.096 |
5.946 ± 0.23 | 7.951 ± 0.579 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.436 ± 0.865 | 3.725 ± 0.287 |
7.02 ± 1.635 | 3.653 ± 0.093 |
8.309 ± 0.723 | 8.453 ± 0.767 |
6.375 ± 0.685 | 6.304 ± 0.53 |
1.146 ± 0.313 | 2.722 ± 0.929 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
