
Flavobacteriaceae bacterium R33
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; unclassified Flavobacteriaceae
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
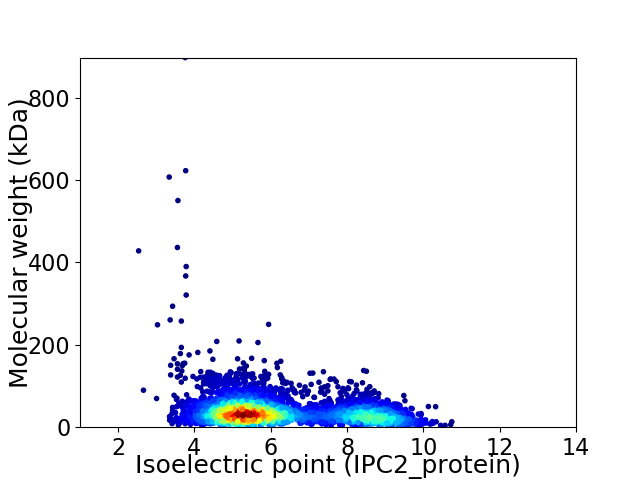
Virtual 2D-PAGE plot for 4113 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6L9E6X2|A0A6L9E6X2_9FLAO Rrf2 family transcriptional regulator OS=Flavobacteriaceae bacterium R33 OX=2697053 GN=GTQ38_00685 PE=4 SV=1
MM1 pKa = 7.71LFLCLWITSDD11 pKa = 5.72LFAQSEE17 pKa = 5.26FITTWDD23 pKa = 3.35TTKK26 pKa = 10.52PGQSNDD32 pKa = 3.09NSITIPAYY40 pKa = 8.84GTYY43 pKa = 10.54DD44 pKa = 3.14VDD46 pKa = 4.17LGNDD50 pKa = 3.36GSYY53 pKa = 11.09EE54 pKa = 4.09LLSQTGTITVDD65 pKa = 3.39VTTHH69 pKa = 6.47NYY71 pKa = 7.39TAGLIQLALRR81 pKa = 11.84NAVSGNRR88 pKa = 11.84LTRR91 pKa = 11.84IHH93 pKa = 6.75FNYY96 pKa = 10.66AGDD99 pKa = 3.85RR100 pKa = 11.84QKK102 pKa = 11.35LLSVDD107 pKa = 3.1QWGSGIYY114 pKa = 9.2WSTMEE119 pKa = 5.07DD120 pKa = 3.28AFSGCTNLEE129 pKa = 3.96VKK131 pKa = 9.67ATDD134 pKa = 3.86FPNLGSVSNMSWMFGGCTSLTGATDD159 pKa = 4.08FSKK162 pKa = 10.65WDD164 pKa = 3.53TGKK167 pKa = 8.68VTDD170 pKa = 4.16MSFMFHH176 pKa = 6.77GAGAFNGNIASWNTDD191 pKa = 2.28KK192 pKa = 10.51VTNMQALFMDD202 pKa = 4.15TSAFNQNIASWNTSNVTTMANMFRR226 pKa = 11.84NAAVFDD232 pKa = 3.89QDD234 pKa = 3.08IGSWDD239 pKa = 3.64TGSVTDD245 pKa = 4.17MVSMFNSASNFDD257 pKa = 3.65QDD259 pKa = 5.28LGDD262 pKa = 3.55WDD264 pKa = 4.85LGKK267 pKa = 10.71LSNGTTMFKK276 pKa = 10.67SSGLSVGNWDD286 pKa = 3.22ATLIGWYY293 pKa = 7.15TQSFTNTPTIGASGLVYY310 pKa = 9.49CTAGTQRR317 pKa = 11.84AALTLNITGDD327 pKa = 3.8SAEE330 pKa = 4.4TTPPTAACQTAVTLQLGTSGTVMLDD355 pKa = 3.0AASVDD360 pKa = 3.54NGSSDD365 pKa = 3.27TCGNVSLGLSQSSFTGSDD383 pKa = 2.8LGTNTVTLTVTDD395 pKa = 4.07PNGNKK400 pKa = 9.67NSCTIVVTVEE410 pKa = 4.26DD411 pKa = 4.92KK412 pKa = 10.78IAPTAACKK420 pKa = 8.76DD421 pKa = 3.74TKK423 pKa = 11.1VEE425 pKa = 3.98LDD427 pKa = 3.53SAGSATITAANVNNNSSDD445 pKa = 3.38NSGSVILSLNTSGYY459 pKa = 9.94DD460 pKa = 3.39CSKK463 pKa = 9.66TGEE466 pKa = 4.19NTVTLTVSDD475 pKa = 4.13GSGNSDD481 pKa = 3.12SCTAKK486 pKa = 9.47VTVEE490 pKa = 4.4DD491 pKa = 4.14KK492 pKa = 10.72LAPSATDD499 pKa = 3.87LAPMNLQCSSVAPAADD515 pKa = 3.06ISIVKK520 pKa = 10.18DD521 pKa = 3.32VTDD524 pKa = 3.66NCTANPTVAFVSDD537 pKa = 3.76VSDD540 pKa = 3.89GNSNPEE546 pKa = 4.24VITRR550 pKa = 11.84TYY552 pKa = 11.1SVTDD556 pKa = 3.51GAGNSTTLTQKK567 pKa = 8.8ITLGDD572 pKa = 3.63TTAPTANMLSPISVEE587 pKa = 4.19CKK589 pKa = 10.32SNVPAADD596 pKa = 3.18IALVTGVADD605 pKa = 3.44NCTANPTVAFVSDD618 pKa = 3.76VSDD621 pKa = 3.89GNSNPEE627 pKa = 4.24VITRR631 pKa = 11.84TYY633 pKa = 11.1SVTDD637 pKa = 3.51GAGNSTPLTQTITVNDD653 pKa = 3.83TTAPTVLCQSVTLQLGTDD671 pKa = 4.08GTASLTANLVDD682 pKa = 3.99NGSNDD687 pKa = 3.08ACGSVTLNLSSYY699 pKa = 11.42SFTTSDD705 pKa = 4.15LGDD708 pKa = 3.44NTVTLTVTDD717 pKa = 4.41PNGNTNTCQATVTIVDD733 pKa = 3.85PASVFVTTWDD743 pKa = 3.3TTKK746 pKa = 10.65PGTSDD751 pKa = 3.22GKK753 pKa = 11.04SITIPVSGTYY763 pKa = 10.4DD764 pKa = 3.04VDD766 pKa = 3.92LGNDD770 pKa = 3.55GTYY773 pKa = 11.09EE774 pKa = 4.28LLDD777 pKa = 3.61QNGTLTLDD785 pKa = 3.24VTTYY789 pKa = 10.42KK790 pKa = 9.35HH791 pKa = 5.21TAGEE795 pKa = 3.97IQLALRR801 pKa = 11.84NAASGNGSLSRR812 pKa = 11.84IHH814 pKa = 6.58FNNSGDD820 pKa = 3.79RR821 pKa = 11.84QKK823 pKa = 11.37LLSVDD828 pKa = 3.12QWGSSITWSTMEE840 pKa = 3.86EE841 pKa = 3.99AFYY844 pKa = 11.13GCANLEE850 pKa = 4.32VKK852 pKa = 9.63ATDD855 pKa = 3.95APNLSYY861 pKa = 9.88VTTMIEE867 pKa = 4.23SFMLCTSLKK876 pKa = 9.1GTTSFPSWNTTGVTDD891 pKa = 3.48MSYY894 pKa = 9.81MFNQASSFNGNISSWEE910 pKa = 4.11TGNVTNMAAMFTGASSFDD928 pKa = 3.86RR929 pKa = 11.84NIGSWNTSGVTNMASMFYY947 pKa = 9.9QASSFNANIGSWNTSNVTTMVSMFYY972 pKa = 9.9QASSFDD978 pKa = 3.88EE979 pKa = 5.59DD980 pKa = 4.25LGNWDD985 pKa = 4.46IGQLTNGTGMLNGSGLSMEE1004 pKa = 4.54NWDD1007 pKa = 3.69DD1008 pKa = 3.9TLIGWHH1014 pKa = 5.64SQGFTNAVTIGASGLHH1030 pKa = 5.55YY1031 pKa = 10.19CQAGAEE1037 pKa = 4.07RR1038 pKa = 11.84AAMTLSFSGDD1048 pKa = 3.72SVEE1051 pKa = 4.53DD1052 pKa = 3.7TAPTASNPASVSVKK1066 pKa = 10.23CSSDD1070 pKa = 3.31VPAADD1075 pKa = 2.91IAVVTDD1081 pKa = 3.68EE1082 pKa = 5.16SDD1084 pKa = 3.2NCTASPTVTFVSDD1097 pKa = 3.62VSDD1100 pKa = 3.92GNSNPEE1106 pKa = 4.23VITRR1110 pKa = 11.84TYY1112 pKa = 10.63RR1113 pKa = 11.84ITDD1116 pKa = 3.34EE1117 pKa = 5.0AGNSTDD1123 pKa = 3.14VTQTITIEE1131 pKa = 4.12DD1132 pKa = 3.74TT1133 pKa = 3.49
MM1 pKa = 7.71LFLCLWITSDD11 pKa = 5.72LFAQSEE17 pKa = 5.26FITTWDD23 pKa = 3.35TTKK26 pKa = 10.52PGQSNDD32 pKa = 3.09NSITIPAYY40 pKa = 8.84GTYY43 pKa = 10.54DD44 pKa = 3.14VDD46 pKa = 4.17LGNDD50 pKa = 3.36GSYY53 pKa = 11.09EE54 pKa = 4.09LLSQTGTITVDD65 pKa = 3.39VTTHH69 pKa = 6.47NYY71 pKa = 7.39TAGLIQLALRR81 pKa = 11.84NAVSGNRR88 pKa = 11.84LTRR91 pKa = 11.84IHH93 pKa = 6.75FNYY96 pKa = 10.66AGDD99 pKa = 3.85RR100 pKa = 11.84QKK102 pKa = 11.35LLSVDD107 pKa = 3.1QWGSGIYY114 pKa = 9.2WSTMEE119 pKa = 5.07DD120 pKa = 3.28AFSGCTNLEE129 pKa = 3.96VKK131 pKa = 9.67ATDD134 pKa = 3.86FPNLGSVSNMSWMFGGCTSLTGATDD159 pKa = 4.08FSKK162 pKa = 10.65WDD164 pKa = 3.53TGKK167 pKa = 8.68VTDD170 pKa = 4.16MSFMFHH176 pKa = 6.77GAGAFNGNIASWNTDD191 pKa = 2.28KK192 pKa = 10.51VTNMQALFMDD202 pKa = 4.15TSAFNQNIASWNTSNVTTMANMFRR226 pKa = 11.84NAAVFDD232 pKa = 3.89QDD234 pKa = 3.08IGSWDD239 pKa = 3.64TGSVTDD245 pKa = 4.17MVSMFNSASNFDD257 pKa = 3.65QDD259 pKa = 5.28LGDD262 pKa = 3.55WDD264 pKa = 4.85LGKK267 pKa = 10.71LSNGTTMFKK276 pKa = 10.67SSGLSVGNWDD286 pKa = 3.22ATLIGWYY293 pKa = 7.15TQSFTNTPTIGASGLVYY310 pKa = 9.49CTAGTQRR317 pKa = 11.84AALTLNITGDD327 pKa = 3.8SAEE330 pKa = 4.4TTPPTAACQTAVTLQLGTSGTVMLDD355 pKa = 3.0AASVDD360 pKa = 3.54NGSSDD365 pKa = 3.27TCGNVSLGLSQSSFTGSDD383 pKa = 2.8LGTNTVTLTVTDD395 pKa = 4.07PNGNKK400 pKa = 9.67NSCTIVVTVEE410 pKa = 4.26DD411 pKa = 4.92KK412 pKa = 10.78IAPTAACKK420 pKa = 8.76DD421 pKa = 3.74TKK423 pKa = 11.1VEE425 pKa = 3.98LDD427 pKa = 3.53SAGSATITAANVNNNSSDD445 pKa = 3.38NSGSVILSLNTSGYY459 pKa = 9.94DD460 pKa = 3.39CSKK463 pKa = 9.66TGEE466 pKa = 4.19NTVTLTVSDD475 pKa = 4.13GSGNSDD481 pKa = 3.12SCTAKK486 pKa = 9.47VTVEE490 pKa = 4.4DD491 pKa = 4.14KK492 pKa = 10.72LAPSATDD499 pKa = 3.87LAPMNLQCSSVAPAADD515 pKa = 3.06ISIVKK520 pKa = 10.18DD521 pKa = 3.32VTDD524 pKa = 3.66NCTANPTVAFVSDD537 pKa = 3.76VSDD540 pKa = 3.89GNSNPEE546 pKa = 4.24VITRR550 pKa = 11.84TYY552 pKa = 11.1SVTDD556 pKa = 3.51GAGNSTTLTQKK567 pKa = 8.8ITLGDD572 pKa = 3.63TTAPTANMLSPISVEE587 pKa = 4.19CKK589 pKa = 10.32SNVPAADD596 pKa = 3.18IALVTGVADD605 pKa = 3.44NCTANPTVAFVSDD618 pKa = 3.76VSDD621 pKa = 3.89GNSNPEE627 pKa = 4.24VITRR631 pKa = 11.84TYY633 pKa = 11.1SVTDD637 pKa = 3.51GAGNSTPLTQTITVNDD653 pKa = 3.83TTAPTVLCQSVTLQLGTDD671 pKa = 4.08GTASLTANLVDD682 pKa = 3.99NGSNDD687 pKa = 3.08ACGSVTLNLSSYY699 pKa = 11.42SFTTSDD705 pKa = 4.15LGDD708 pKa = 3.44NTVTLTVTDD717 pKa = 4.41PNGNTNTCQATVTIVDD733 pKa = 3.85PASVFVTTWDD743 pKa = 3.3TTKK746 pKa = 10.65PGTSDD751 pKa = 3.22GKK753 pKa = 11.04SITIPVSGTYY763 pKa = 10.4DD764 pKa = 3.04VDD766 pKa = 3.92LGNDD770 pKa = 3.55GTYY773 pKa = 11.09EE774 pKa = 4.28LLDD777 pKa = 3.61QNGTLTLDD785 pKa = 3.24VTTYY789 pKa = 10.42KK790 pKa = 9.35HH791 pKa = 5.21TAGEE795 pKa = 3.97IQLALRR801 pKa = 11.84NAASGNGSLSRR812 pKa = 11.84IHH814 pKa = 6.58FNNSGDD820 pKa = 3.79RR821 pKa = 11.84QKK823 pKa = 11.37LLSVDD828 pKa = 3.12QWGSSITWSTMEE840 pKa = 3.86EE841 pKa = 3.99AFYY844 pKa = 11.13GCANLEE850 pKa = 4.32VKK852 pKa = 9.63ATDD855 pKa = 3.95APNLSYY861 pKa = 9.88VTTMIEE867 pKa = 4.23SFMLCTSLKK876 pKa = 9.1GTTSFPSWNTTGVTDD891 pKa = 3.48MSYY894 pKa = 9.81MFNQASSFNGNISSWEE910 pKa = 4.11TGNVTNMAAMFTGASSFDD928 pKa = 3.86RR929 pKa = 11.84NIGSWNTSGVTNMASMFYY947 pKa = 9.9QASSFNANIGSWNTSNVTTMVSMFYY972 pKa = 9.9QASSFDD978 pKa = 3.88EE979 pKa = 5.59DD980 pKa = 4.25LGNWDD985 pKa = 4.46IGQLTNGTGMLNGSGLSMEE1004 pKa = 4.54NWDD1007 pKa = 3.69DD1008 pKa = 3.9TLIGWHH1014 pKa = 5.64SQGFTNAVTIGASGLHH1030 pKa = 5.55YY1031 pKa = 10.19CQAGAEE1037 pKa = 4.07RR1038 pKa = 11.84AAMTLSFSGDD1048 pKa = 3.72SVEE1051 pKa = 4.53DD1052 pKa = 3.7TAPTASNPASVSVKK1066 pKa = 10.23CSSDD1070 pKa = 3.31VPAADD1075 pKa = 2.91IAVVTDD1081 pKa = 3.68EE1082 pKa = 5.16SDD1084 pKa = 3.2NCTASPTVTFVSDD1097 pKa = 3.62VSDD1100 pKa = 3.92GNSNPEE1106 pKa = 4.23VITRR1110 pKa = 11.84TYY1112 pKa = 10.63RR1113 pKa = 11.84ITDD1116 pKa = 3.34EE1117 pKa = 5.0AGNSTDD1123 pKa = 3.14VTQTITIEE1131 pKa = 4.12DD1132 pKa = 3.74TT1133 pKa = 3.49
Molecular weight: 118.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6L9EHV2|A0A6L9EHV2_9FLAO MMPL family transporter OS=Flavobacteriaceae bacterium R33 OX=2697053 GN=GTQ38_19655 PE=4 SV=1
MM1 pKa = 7.74PGYY4 pKa = 9.93IKK6 pKa = 10.67NRR8 pKa = 11.84IKK10 pKa = 11.03GRR12 pKa = 11.84GFPWFRR18 pKa = 11.84AVGMICLFPIRR29 pKa = 11.84HH30 pKa = 6.45KK31 pKa = 10.84GAQCAWAGRR40 pKa = 11.84YY41 pKa = 8.33MDD43 pKa = 5.56DD44 pKa = 2.72MHH46 pKa = 9.12LLFRR50 pKa = 11.84VLKK53 pKa = 10.05RR54 pKa = 11.84SKK56 pKa = 9.58PLPSAWEE63 pKa = 3.87VLGG66 pKa = 4.16
MM1 pKa = 7.74PGYY4 pKa = 9.93IKK6 pKa = 10.67NRR8 pKa = 11.84IKK10 pKa = 11.03GRR12 pKa = 11.84GFPWFRR18 pKa = 11.84AVGMICLFPIRR29 pKa = 11.84HH30 pKa = 6.45KK31 pKa = 10.84GAQCAWAGRR40 pKa = 11.84YY41 pKa = 8.33MDD43 pKa = 5.56DD44 pKa = 2.72MHH46 pKa = 9.12LLFRR50 pKa = 11.84VLKK53 pKa = 10.05RR54 pKa = 11.84SKK56 pKa = 9.58PLPSAWEE63 pKa = 3.87VLGG66 pKa = 4.16
Molecular weight: 7.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1474473 |
32 |
8468 |
358.5 |
40.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.943 ± 0.037 | 0.766 ± 0.016 |
5.927 ± 0.048 | 6.8 ± 0.06 |
5.036 ± 0.036 | 7.095 ± 0.044 |
1.798 ± 0.021 | 6.905 ± 0.04 |
6.326 ± 0.076 | 9.66 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.223 ± 0.021 | 5.243 ± 0.047 |
3.75 ± 0.032 | 3.509 ± 0.023 |
4.236 ± 0.04 | 6.704 ± 0.052 |
5.693 ± 0.103 | 6.293 ± 0.032 |
1.227 ± 0.017 | 3.865 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
