
Mesorhizobium alhagi CCNWXJ12-2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Mesorhizobium; Mesorhizobium alhagi
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
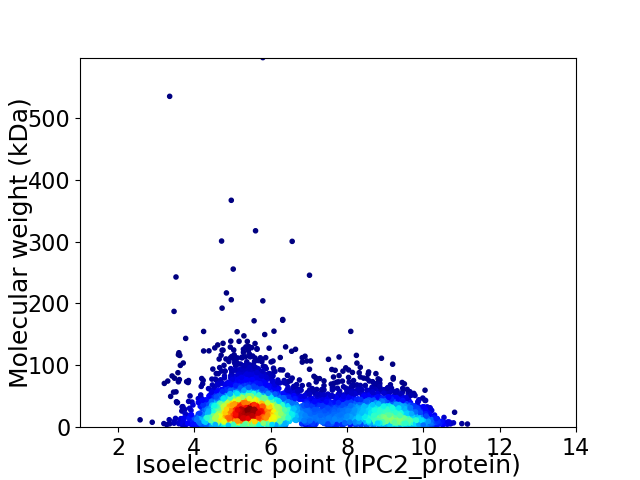
Virtual 2D-PAGE plot for 7195 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
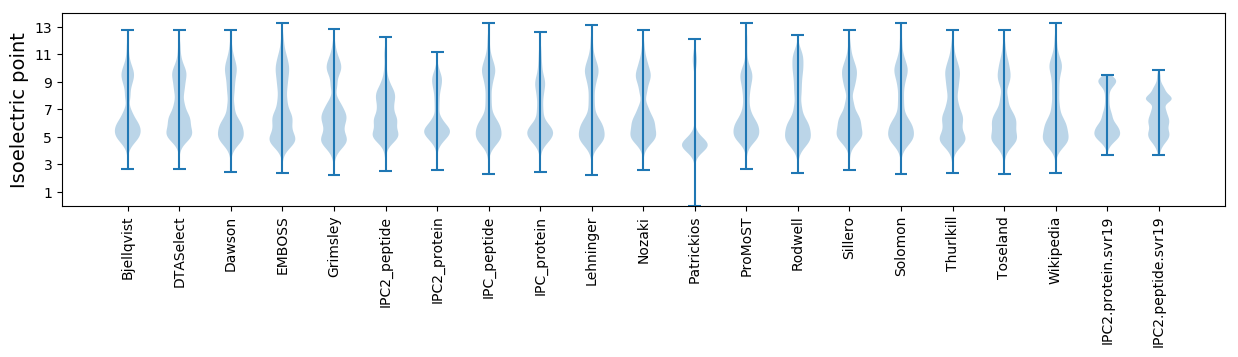
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H0HTG2|H0HTG2_9RHIZ Peptidase M24 OS=Mesorhizobium alhagi CCNWXJ12-2 OX=1107882 GN=MAXJ12_17268 PE=3 SV=1
MM1 pKa = 6.81TTTITSNLTTQFNITAPNDD20 pKa = 3.06NWVLANGVSINVANGHH36 pKa = 6.45GIFNGGAYY44 pKa = 9.38HH45 pKa = 7.32DD46 pKa = 4.3SVVEE50 pKa = 4.05VRR52 pKa = 11.84GHH54 pKa = 4.82ITANATYY61 pKa = 10.16KK62 pKa = 10.78AGVYY66 pKa = 10.23SDD68 pKa = 4.24GLDD71 pKa = 4.19AILSVADD78 pKa = 4.4LGTIHH83 pKa = 7.68AYY85 pKa = 10.74NGMAFYY91 pKa = 11.22GDD93 pKa = 3.22GTAADD98 pKa = 3.42NHH100 pKa = 6.59GIITASNIGMYY111 pKa = 10.99GNGNDD116 pKa = 3.54IKK118 pKa = 10.54ISNFGSIVAEE128 pKa = 3.65AGIYY132 pKa = 9.92VEE134 pKa = 4.57AASNHH139 pKa = 4.84SQIEE143 pKa = 4.26NNGKK147 pKa = 9.04IEE149 pKa = 4.56GYY151 pKa = 10.26SGIISVNSRR160 pKa = 11.84LDD162 pKa = 3.12IKK164 pKa = 10.89LGSQSVIDD172 pKa = 3.75VQGIGIISQGQLTTYY187 pKa = 9.88VEE189 pKa = 4.16NYY191 pKa = 10.62GFIKK195 pKa = 10.36SANLAIKK202 pKa = 10.45GGDD205 pKa = 3.6GEE207 pKa = 5.67DD208 pKa = 3.14IVINYY213 pKa = 9.8GNIQGGIFLGGGNDD227 pKa = 3.5FFDD230 pKa = 4.02NRR232 pKa = 11.84TGTIDD237 pKa = 3.39HH238 pKa = 6.76AVDD241 pKa = 3.53GGLGDD246 pKa = 4.06DD247 pKa = 4.36YY248 pKa = 11.6YY249 pKa = 12.06VFGAKK254 pKa = 9.83PFIIIDD260 pKa = 3.45SGGNDD265 pKa = 3.75TIGTEE270 pKa = 4.05ITRR273 pKa = 11.84SLASYY278 pKa = 11.08GNIEE282 pKa = 4.24NLHH285 pKa = 6.18LWGSANINGTGNQLANVISGNSGNNILDD313 pKa = 3.8GGADD317 pKa = 3.62NIVDD321 pKa = 3.55QLEE324 pKa = 4.15GRR326 pKa = 11.84EE327 pKa = 4.24GNDD330 pKa = 3.24TYY332 pKa = 11.93VLGAGFDD339 pKa = 3.61IVIEE343 pKa = 4.54SIFPSAAFVGTVYY356 pKa = 10.9GGVDD360 pKa = 4.31TITSTISRR368 pKa = 11.84SLGDD372 pKa = 3.55YY373 pKa = 11.07AHH375 pKa = 6.61VEE377 pKa = 3.99NLKK380 pKa = 10.9LLGSANINGTGDD392 pKa = 3.1EE393 pKa = 4.28AANVITGNSGNNILNGGGGNDD414 pKa = 3.93TLNGGAGADD423 pKa = 3.41AMFGGTGDD431 pKa = 4.79DD432 pKa = 3.41IYY434 pKa = 11.01IVAAAGDD441 pKa = 4.0TTIEE445 pKa = 4.12NAGEE449 pKa = 4.08GTDD452 pKa = 3.6TVRR455 pKa = 11.84SYY457 pKa = 11.42IDD459 pKa = 3.02WVLANNVEE467 pKa = 4.08RR468 pKa = 11.84LEE470 pKa = 4.15LQGSGNLNGTGNALNNTLVGNSGNNLLNGGAGNDD504 pKa = 3.64YY505 pKa = 10.05MVGGGGDD512 pKa = 3.42DD513 pKa = 3.72TFIVAAAGDD522 pKa = 3.86ITVEE526 pKa = 4.21SAGGGTDD533 pKa = 3.27TVRR536 pKa = 11.84SYY538 pKa = 11.69INWTLGANVEE548 pKa = 4.09RR549 pKa = 11.84LEE551 pKa = 4.21LQGSSNLNGTGNSLNNTLVGNSGNNSLSGGDD582 pKa = 3.49GNDD585 pKa = 3.95YY586 pKa = 9.73ITGGAGNDD594 pKa = 3.61TLTGGVGNDD603 pKa = 3.67TLIGGAGSDD612 pKa = 4.11SLNGGAGNDD621 pKa = 3.17RR622 pKa = 11.84FDD624 pKa = 4.56FDD626 pKa = 5.06LVSHH630 pKa = 6.42SPVGPALRR638 pKa = 11.84DD639 pKa = 3.76SIVGGFSHH647 pKa = 6.99GFDD650 pKa = 5.2LIDD653 pKa = 5.12LATIDD658 pKa = 4.92ANTLVAGNQAFSFIGSAAFSGAAGQLRR685 pKa = 11.84YY686 pKa = 8.45TNYY689 pKa = 10.23NGTVIIDD696 pKa = 3.49ADD698 pKa = 3.99VNGDD702 pKa = 4.38SIADD706 pKa = 3.77MQILVAGTSFMTGTDD721 pKa = 3.75FVLL724 pKa = 4.53
MM1 pKa = 6.81TTTITSNLTTQFNITAPNDD20 pKa = 3.06NWVLANGVSINVANGHH36 pKa = 6.45GIFNGGAYY44 pKa = 9.38HH45 pKa = 7.32DD46 pKa = 4.3SVVEE50 pKa = 4.05VRR52 pKa = 11.84GHH54 pKa = 4.82ITANATYY61 pKa = 10.16KK62 pKa = 10.78AGVYY66 pKa = 10.23SDD68 pKa = 4.24GLDD71 pKa = 4.19AILSVADD78 pKa = 4.4LGTIHH83 pKa = 7.68AYY85 pKa = 10.74NGMAFYY91 pKa = 11.22GDD93 pKa = 3.22GTAADD98 pKa = 3.42NHH100 pKa = 6.59GIITASNIGMYY111 pKa = 10.99GNGNDD116 pKa = 3.54IKK118 pKa = 10.54ISNFGSIVAEE128 pKa = 3.65AGIYY132 pKa = 9.92VEE134 pKa = 4.57AASNHH139 pKa = 4.84SQIEE143 pKa = 4.26NNGKK147 pKa = 9.04IEE149 pKa = 4.56GYY151 pKa = 10.26SGIISVNSRR160 pKa = 11.84LDD162 pKa = 3.12IKK164 pKa = 10.89LGSQSVIDD172 pKa = 3.75VQGIGIISQGQLTTYY187 pKa = 9.88VEE189 pKa = 4.16NYY191 pKa = 10.62GFIKK195 pKa = 10.36SANLAIKK202 pKa = 10.45GGDD205 pKa = 3.6GEE207 pKa = 5.67DD208 pKa = 3.14IVINYY213 pKa = 9.8GNIQGGIFLGGGNDD227 pKa = 3.5FFDD230 pKa = 4.02NRR232 pKa = 11.84TGTIDD237 pKa = 3.39HH238 pKa = 6.76AVDD241 pKa = 3.53GGLGDD246 pKa = 4.06DD247 pKa = 4.36YY248 pKa = 11.6YY249 pKa = 12.06VFGAKK254 pKa = 9.83PFIIIDD260 pKa = 3.45SGGNDD265 pKa = 3.75TIGTEE270 pKa = 4.05ITRR273 pKa = 11.84SLASYY278 pKa = 11.08GNIEE282 pKa = 4.24NLHH285 pKa = 6.18LWGSANINGTGNQLANVISGNSGNNILDD313 pKa = 3.8GGADD317 pKa = 3.62NIVDD321 pKa = 3.55QLEE324 pKa = 4.15GRR326 pKa = 11.84EE327 pKa = 4.24GNDD330 pKa = 3.24TYY332 pKa = 11.93VLGAGFDD339 pKa = 3.61IVIEE343 pKa = 4.54SIFPSAAFVGTVYY356 pKa = 10.9GGVDD360 pKa = 4.31TITSTISRR368 pKa = 11.84SLGDD372 pKa = 3.55YY373 pKa = 11.07AHH375 pKa = 6.61VEE377 pKa = 3.99NLKK380 pKa = 10.9LLGSANINGTGDD392 pKa = 3.1EE393 pKa = 4.28AANVITGNSGNNILNGGGGNDD414 pKa = 3.93TLNGGAGADD423 pKa = 3.41AMFGGTGDD431 pKa = 4.79DD432 pKa = 3.41IYY434 pKa = 11.01IVAAAGDD441 pKa = 4.0TTIEE445 pKa = 4.12NAGEE449 pKa = 4.08GTDD452 pKa = 3.6TVRR455 pKa = 11.84SYY457 pKa = 11.42IDD459 pKa = 3.02WVLANNVEE467 pKa = 4.08RR468 pKa = 11.84LEE470 pKa = 4.15LQGSGNLNGTGNALNNTLVGNSGNNLLNGGAGNDD504 pKa = 3.64YY505 pKa = 10.05MVGGGGDD512 pKa = 3.42DD513 pKa = 3.72TFIVAAAGDD522 pKa = 3.86ITVEE526 pKa = 4.21SAGGGTDD533 pKa = 3.27TVRR536 pKa = 11.84SYY538 pKa = 11.69INWTLGANVEE548 pKa = 4.09RR549 pKa = 11.84LEE551 pKa = 4.21LQGSSNLNGTGNSLNNTLVGNSGNNSLSGGDD582 pKa = 3.49GNDD585 pKa = 3.95YY586 pKa = 9.73ITGGAGNDD594 pKa = 3.61TLTGGVGNDD603 pKa = 3.67TLIGGAGSDD612 pKa = 4.11SLNGGAGNDD621 pKa = 3.17RR622 pKa = 11.84FDD624 pKa = 4.56FDD626 pKa = 5.06LVSHH630 pKa = 6.42SPVGPALRR638 pKa = 11.84DD639 pKa = 3.76SIVGGFSHH647 pKa = 6.99GFDD650 pKa = 5.2LIDD653 pKa = 5.12LATIDD658 pKa = 4.92ANTLVAGNQAFSFIGSAAFSGAAGQLRR685 pKa = 11.84YY686 pKa = 8.45TNYY689 pKa = 10.23NGTVIIDD696 pKa = 3.49ADD698 pKa = 3.99VNGDD702 pKa = 4.38SIADD706 pKa = 3.77MQILVAGTSFMTGTDD721 pKa = 3.75FVLL724 pKa = 4.53
Molecular weight: 73.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H0HNS7|H0HNS7_9RHIZ Uncharacterized protein OS=Mesorhizobium alhagi CCNWXJ12-2 OX=1107882 GN=MAXJ12_09011 PE=4 SV=1
MM1 pKa = 7.48IAPVRR6 pKa = 11.84LPLRR10 pKa = 11.84VAPGHH15 pKa = 5.71RR16 pKa = 11.84QAVTAPQQAKK26 pKa = 8.75TIWLVRR32 pKa = 11.84MFSPNFATFPLVGRR46 pKa = 11.84RR47 pKa = 11.84QALRR51 pKa = 3.46
MM1 pKa = 7.48IAPVRR6 pKa = 11.84LPLRR10 pKa = 11.84VAPGHH15 pKa = 5.71RR16 pKa = 11.84QAVTAPQQAKK26 pKa = 8.75TIWLVRR32 pKa = 11.84MFSPNFATFPLVGRR46 pKa = 11.84RR47 pKa = 11.84QALRR51 pKa = 3.46
Molecular weight: 5.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1987422 |
19 |
5515 |
276.2 |
30.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.38 ± 0.039 | 0.83 ± 0.011 |
5.67 ± 0.026 | 5.996 ± 0.03 |
3.919 ± 0.017 | 8.603 ± 0.043 |
2.011 ± 0.016 | 5.421 ± 0.024 |
3.465 ± 0.03 | 9.789 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.538 ± 0.016 | 2.712 ± 0.023 |
5.071 ± 0.025 | 2.958 ± 0.019 |
7.068 ± 0.033 | 5.553 ± 0.022 |
5.156 ± 0.033 | 7.317 ± 0.022 |
1.353 ± 0.012 | 2.192 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
