
Commensalibacter sp. MX01
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Commensalibacter; unclassified Commensalibacter
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
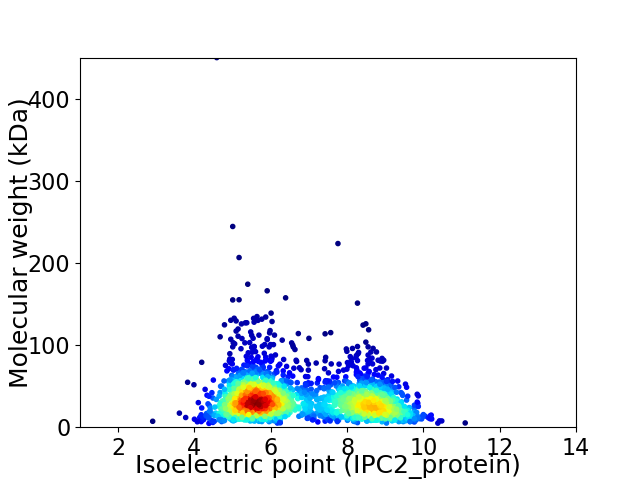
Virtual 2D-PAGE plot for 2060 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W7DYL5|W7DYL5_9PROT AFG1 family ATPase OS=Commensalibacter sp. MX01 OX=1208583 GN=COMX_07290 PE=4 SV=1
MM1 pKa = 8.13DD2 pKa = 5.36NNDD5 pKa = 3.36ANEE8 pKa = 4.29NYY10 pKa = 10.08VFDD13 pKa = 5.64PDD15 pKa = 4.33SEE17 pKa = 4.86SVDD20 pKa = 3.18PDD22 pKa = 3.31QPFEE26 pKa = 4.19VGSDD30 pKa = 3.59FFDD33 pKa = 3.77NPITKK38 pKa = 9.54EE39 pKa = 3.76ASVKK43 pKa = 10.46FFGDD47 pKa = 2.97LRR49 pKa = 11.84DD50 pKa = 4.18YY51 pKa = 9.9ITSGNVKK58 pKa = 10.39DD59 pKa = 5.1VVLEE63 pKa = 4.2PNTHH67 pKa = 5.93IDD69 pKa = 4.03ANEE72 pKa = 3.92LDD74 pKa = 3.93YY75 pKa = 11.52TPIVTMYY82 pKa = 10.68SFSDD86 pKa = 3.63SQNIFYY92 pKa = 10.76LSTGKK97 pKa = 10.24FPQDD101 pKa = 2.75TEE103 pKa = 4.31YY104 pKa = 11.65AEE106 pKa = 4.34VTLGPGQTTDD116 pKa = 4.06NINGIPKK123 pKa = 9.76IQGDD127 pKa = 4.52GNNPNYY133 pKa = 10.27VRR135 pKa = 11.84IIGGVDD141 pKa = 3.52FNYY144 pKa = 10.3IADD147 pKa = 4.21GEE149 pKa = 4.43SGEE152 pKa = 5.05LISNNPLYY160 pKa = 10.54FYY162 pKa = 11.37GRR164 pKa = 11.84GLDD167 pKa = 3.67GNTSHH172 pKa = 6.6MGNWTIYY179 pKa = 9.98TDD181 pKa = 4.74DD182 pKa = 4.69FVDD185 pKa = 4.28PDD187 pKa = 3.5TGEE190 pKa = 4.37IYY192 pKa = 10.85DD193 pKa = 4.59DD194 pKa = 5.35DD195 pKa = 6.29RR196 pKa = 11.84DD197 pKa = 4.01TMIQTTDD204 pKa = 3.28GNNKK208 pKa = 7.94ITTGYY213 pKa = 10.49ARR215 pKa = 11.84STIDD219 pKa = 4.11LGSGNNIVYY228 pKa = 10.24SQGHH232 pKa = 5.08DD233 pKa = 3.52TITSSRR239 pKa = 11.84GGYY242 pKa = 7.18QTVTLTGSQSKK253 pKa = 8.33LTMGGNTTILDD264 pKa = 3.34IGNNNNISVGKK275 pKa = 9.9NSSVYY280 pKa = 10.26GAASDD285 pKa = 3.66IVQFDD290 pKa = 3.49TGDD293 pKa = 3.61MNEE296 pKa = 4.06YY297 pKa = 9.16TAGNNSTISATNGTDD312 pKa = 3.04LKK314 pKa = 10.97VIHH317 pKa = 6.59GLDD320 pKa = 2.92NTYY323 pKa = 10.93NINHH327 pKa = 6.76NLTFLNGSGNTVINIKK343 pKa = 9.05GTLAAFGAGSSQYY356 pKa = 10.08TLNAEE361 pKa = 4.71NIDD364 pKa = 3.98STSGSIFVANEE375 pKa = 3.63GNEE378 pKa = 4.23TLDD381 pKa = 4.04AASSSANLQIWANTVAGANNLLAKK405 pKa = 10.31AGQGDD410 pKa = 3.94DD411 pKa = 3.43TLAAGVGNATFTGGVGNNLFMFTKK435 pKa = 10.41EE436 pKa = 4.12SVQDD440 pKa = 3.46GTTIITDD447 pKa = 3.94FTNNDD452 pKa = 3.27KK453 pKa = 10.8IALYY457 pKa = 10.58NYY459 pKa = 10.17GLGSNGLTQILQQSQNDD476 pKa = 3.77ANGNAVLKK484 pKa = 8.64LTNNAAIVIEE494 pKa = 4.32GVSVSSLSTKK504 pKa = 10.36QFDD507 pKa = 3.56IADD510 pKa = 4.34FSHH513 pKa = 6.85
MM1 pKa = 8.13DD2 pKa = 5.36NNDD5 pKa = 3.36ANEE8 pKa = 4.29NYY10 pKa = 10.08VFDD13 pKa = 5.64PDD15 pKa = 4.33SEE17 pKa = 4.86SVDD20 pKa = 3.18PDD22 pKa = 3.31QPFEE26 pKa = 4.19VGSDD30 pKa = 3.59FFDD33 pKa = 3.77NPITKK38 pKa = 9.54EE39 pKa = 3.76ASVKK43 pKa = 10.46FFGDD47 pKa = 2.97LRR49 pKa = 11.84DD50 pKa = 4.18YY51 pKa = 9.9ITSGNVKK58 pKa = 10.39DD59 pKa = 5.1VVLEE63 pKa = 4.2PNTHH67 pKa = 5.93IDD69 pKa = 4.03ANEE72 pKa = 3.92LDD74 pKa = 3.93YY75 pKa = 11.52TPIVTMYY82 pKa = 10.68SFSDD86 pKa = 3.63SQNIFYY92 pKa = 10.76LSTGKK97 pKa = 10.24FPQDD101 pKa = 2.75TEE103 pKa = 4.31YY104 pKa = 11.65AEE106 pKa = 4.34VTLGPGQTTDD116 pKa = 4.06NINGIPKK123 pKa = 9.76IQGDD127 pKa = 4.52GNNPNYY133 pKa = 10.27VRR135 pKa = 11.84IIGGVDD141 pKa = 3.52FNYY144 pKa = 10.3IADD147 pKa = 4.21GEE149 pKa = 4.43SGEE152 pKa = 5.05LISNNPLYY160 pKa = 10.54FYY162 pKa = 11.37GRR164 pKa = 11.84GLDD167 pKa = 3.67GNTSHH172 pKa = 6.6MGNWTIYY179 pKa = 9.98TDD181 pKa = 4.74DD182 pKa = 4.69FVDD185 pKa = 4.28PDD187 pKa = 3.5TGEE190 pKa = 4.37IYY192 pKa = 10.85DD193 pKa = 4.59DD194 pKa = 5.35DD195 pKa = 6.29RR196 pKa = 11.84DD197 pKa = 4.01TMIQTTDD204 pKa = 3.28GNNKK208 pKa = 7.94ITTGYY213 pKa = 10.49ARR215 pKa = 11.84STIDD219 pKa = 4.11LGSGNNIVYY228 pKa = 10.24SQGHH232 pKa = 5.08DD233 pKa = 3.52TITSSRR239 pKa = 11.84GGYY242 pKa = 7.18QTVTLTGSQSKK253 pKa = 8.33LTMGGNTTILDD264 pKa = 3.34IGNNNNISVGKK275 pKa = 9.9NSSVYY280 pKa = 10.26GAASDD285 pKa = 3.66IVQFDD290 pKa = 3.49TGDD293 pKa = 3.61MNEE296 pKa = 4.06YY297 pKa = 9.16TAGNNSTISATNGTDD312 pKa = 3.04LKK314 pKa = 10.97VIHH317 pKa = 6.59GLDD320 pKa = 2.92NTYY323 pKa = 10.93NINHH327 pKa = 6.76NLTFLNGSGNTVINIKK343 pKa = 9.05GTLAAFGAGSSQYY356 pKa = 10.08TLNAEE361 pKa = 4.71NIDD364 pKa = 3.98STSGSIFVANEE375 pKa = 3.63GNEE378 pKa = 4.23TLDD381 pKa = 4.04AASSSANLQIWANTVAGANNLLAKK405 pKa = 10.31AGQGDD410 pKa = 3.94DD411 pKa = 3.43TLAAGVGNATFTGGVGNNLFMFTKK435 pKa = 10.41EE436 pKa = 4.12SVQDD440 pKa = 3.46GTTIITDD447 pKa = 3.94FTNNDD452 pKa = 3.27KK453 pKa = 10.8IALYY457 pKa = 10.58NYY459 pKa = 10.17GLGSNGLTQILQQSQNDD476 pKa = 3.77ANGNAVLKK484 pKa = 8.64LTNNAAIVIEE494 pKa = 4.32GVSVSSLSTKK504 pKa = 10.36QFDD507 pKa = 3.56IADD510 pKa = 4.34FSHH513 pKa = 6.85
Molecular weight: 54.67 kDa
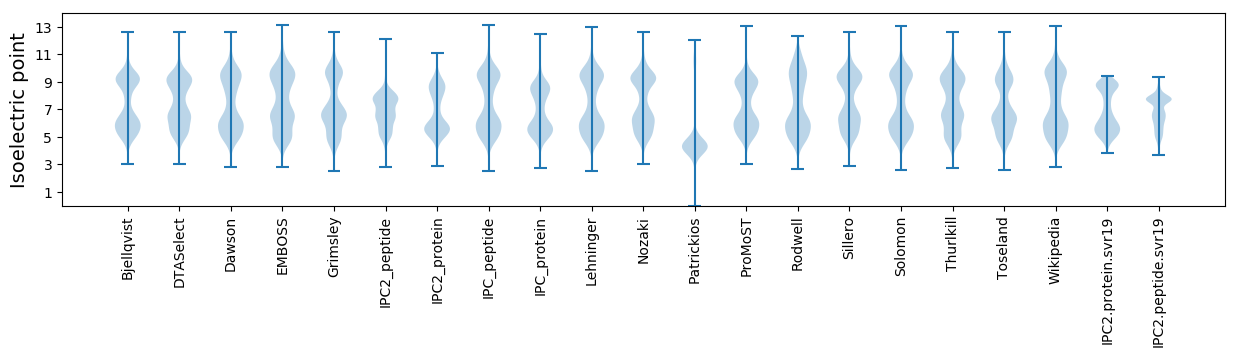
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W7DVK7|W7DVK7_9PROT Methionine aminopeptidase OS=Commensalibacter sp. MX01 OX=1208583 GN=map PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.09QPSKK9 pKa = 9.06IVRR12 pKa = 11.84KK13 pKa = 9.03HH14 pKa = 4.06RR15 pKa = 11.84HH16 pKa = 4.08GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.49VITNRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.87RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.09QPSKK9 pKa = 9.06IVRR12 pKa = 11.84KK13 pKa = 9.03HH14 pKa = 4.06RR15 pKa = 11.84HH16 pKa = 4.08GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.49VITNRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.87RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
686148 |
41 |
4383 |
333.1 |
37.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.028 ± 0.065 | 1.114 ± 0.02 |
5.163 ± 0.041 | 5.422 ± 0.059 |
4.262 ± 0.044 | 6.91 ± 0.071 |
2.27 ± 0.027 | 7.752 ± 0.051 |
5.968 ± 0.045 | 9.948 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.455 ± 0.026 | 4.763 ± 0.076 |
4.38 ± 0.038 | 4.592 ± 0.049 |
4.366 ± 0.044 | 6.389 ± 0.046 |
5.428 ± 0.048 | 6.262 ± 0.04 |
1.292 ± 0.022 | 3.234 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
