
Circoviridae 13 LDMD-2013
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; environmental samples
Average proteome isoelectric point is 7.35
Get precalculated fractions of proteins
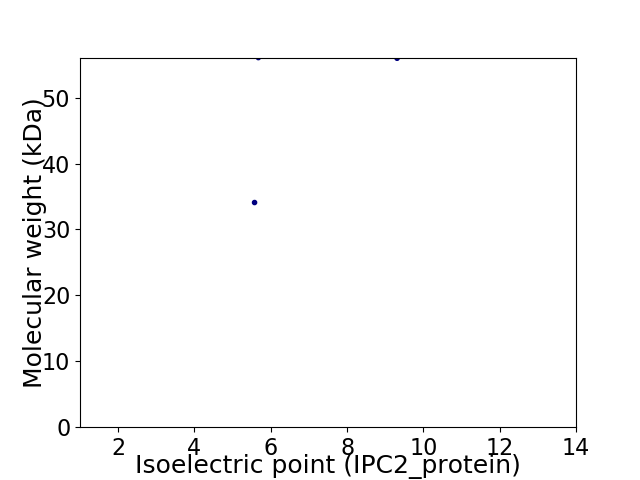
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
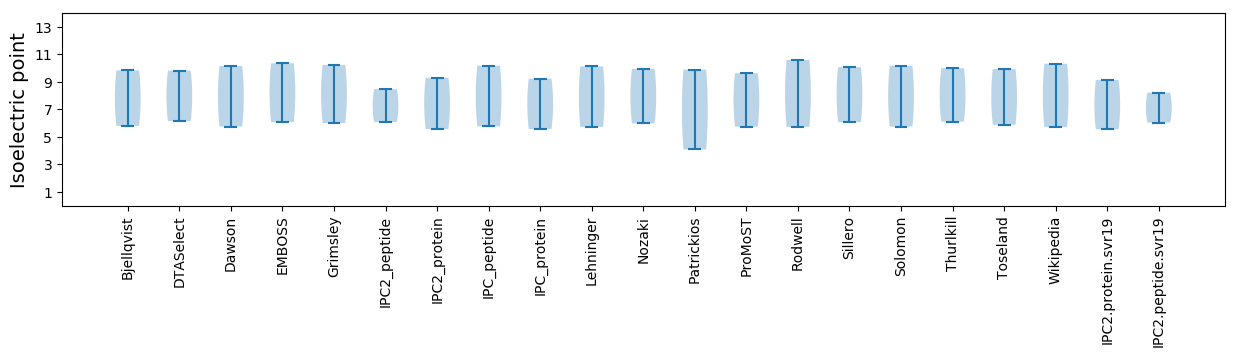
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5T695|S5T695_9CIRC Uncharacterized protein OS=Circoviridae 13 LDMD-2013 OX=1379717 PE=4 SV=1
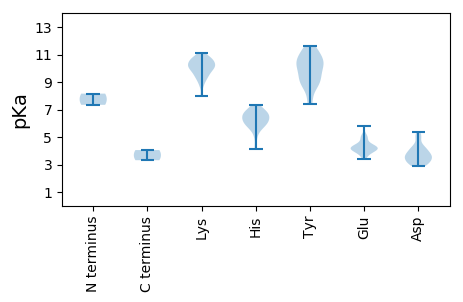
MM1 pKa = 7.34SKK3 pKa = 10.28YY4 pKa = 9.49WCFTLNNYY12 pKa = 7.41TRR14 pKa = 11.84EE15 pKa = 3.94EE16 pKa = 4.14EE17 pKa = 4.2EE18 pKa = 4.06AVKK21 pKa = 10.39ALCEE25 pKa = 4.27GEE27 pKa = 5.0SPIADD32 pKa = 3.34YY33 pKa = 10.94LVYY36 pKa = 10.53GRR38 pKa = 11.84EE39 pKa = 3.93RR40 pKa = 11.84GEE42 pKa = 4.9GEE44 pKa = 4.42DD45 pKa = 3.84TPHH48 pKa = 5.89LQGYY52 pKa = 9.64IEE54 pKa = 4.21FKK56 pKa = 9.34NRR58 pKa = 11.84KK59 pKa = 8.69RR60 pKa = 11.84FSTVRR65 pKa = 11.84TVIPRR70 pKa = 11.84AHH72 pKa = 6.9IEE74 pKa = 3.78RR75 pKa = 11.84RR76 pKa = 11.84MGSASEE82 pKa = 3.6AAAYY86 pKa = 8.56CKK88 pKa = 10.06KK89 pKa = 10.83DD90 pKa = 3.07EE91 pKa = 4.85DD92 pKa = 4.19FFEE95 pKa = 5.81HH96 pKa = 6.74GVISQGQGARR106 pKa = 11.84NDD108 pKa = 3.61LARR111 pKa = 11.84LHH113 pKa = 6.1EE114 pKa = 5.16AIFVEE119 pKa = 4.76QKK121 pKa = 10.54SVAHH125 pKa = 5.12VQEE128 pKa = 5.04EE129 pKa = 5.12YY130 pKa = 10.01FGSYY134 pKa = 8.96IKK136 pKa = 9.34YY137 pKa = 9.28HH138 pKa = 6.31RR139 pKa = 11.84GIEE142 pKa = 4.09KK143 pKa = 10.4VLQQRR148 pKa = 11.84AKK150 pKa = 9.54PRR152 pKa = 11.84TWKK155 pKa = 10.63SRR157 pKa = 11.84VVVYY161 pKa = 8.91WGPTGTGKK169 pKa = 7.96TRR171 pKa = 11.84RR172 pKa = 11.84CFASSDD178 pKa = 3.78PPTWIYY184 pKa = 11.35SSDD187 pKa = 3.03GWFDD191 pKa = 3.99GYY193 pKa = 11.25QGDD196 pKa = 4.09EE197 pKa = 4.15TVLFDD202 pKa = 5.35DD203 pKa = 4.97FGGHH207 pKa = 5.76EE208 pKa = 4.34FRR210 pKa = 11.84LTHH213 pKa = 6.75LLKK216 pKa = 11.12LLDD219 pKa = 4.45RR220 pKa = 11.84YY221 pKa = 10.07PMRR224 pKa = 11.84VKK226 pKa = 10.8VKK228 pKa = 10.06GSFVQWCPKK237 pKa = 9.3TIYY240 pKa = 8.68ITSNIEE246 pKa = 3.47PAFWYY251 pKa = 9.37PNARR255 pKa = 11.84EE256 pKa = 4.05VHH258 pKa = 6.27RR259 pKa = 11.84EE260 pKa = 3.43ALMRR264 pKa = 11.84RR265 pKa = 11.84IDD267 pKa = 5.09DD268 pKa = 3.71IEE270 pKa = 4.07EE271 pKa = 4.17MADD274 pKa = 3.04EE275 pKa = 4.47WVPPVPVEE283 pKa = 4.31NEE285 pKa = 4.23VIDD288 pKa = 4.19LTDD291 pKa = 3.22EE292 pKa = 4.05
MM1 pKa = 7.34SKK3 pKa = 10.28YY4 pKa = 9.49WCFTLNNYY12 pKa = 7.41TRR14 pKa = 11.84EE15 pKa = 3.94EE16 pKa = 4.14EE17 pKa = 4.2EE18 pKa = 4.06AVKK21 pKa = 10.39ALCEE25 pKa = 4.27GEE27 pKa = 5.0SPIADD32 pKa = 3.34YY33 pKa = 10.94LVYY36 pKa = 10.53GRR38 pKa = 11.84EE39 pKa = 3.93RR40 pKa = 11.84GEE42 pKa = 4.9GEE44 pKa = 4.42DD45 pKa = 3.84TPHH48 pKa = 5.89LQGYY52 pKa = 9.64IEE54 pKa = 4.21FKK56 pKa = 9.34NRR58 pKa = 11.84KK59 pKa = 8.69RR60 pKa = 11.84FSTVRR65 pKa = 11.84TVIPRR70 pKa = 11.84AHH72 pKa = 6.9IEE74 pKa = 3.78RR75 pKa = 11.84RR76 pKa = 11.84MGSASEE82 pKa = 3.6AAAYY86 pKa = 8.56CKK88 pKa = 10.06KK89 pKa = 10.83DD90 pKa = 3.07EE91 pKa = 4.85DD92 pKa = 4.19FFEE95 pKa = 5.81HH96 pKa = 6.74GVISQGQGARR106 pKa = 11.84NDD108 pKa = 3.61LARR111 pKa = 11.84LHH113 pKa = 6.1EE114 pKa = 5.16AIFVEE119 pKa = 4.76QKK121 pKa = 10.54SVAHH125 pKa = 5.12VQEE128 pKa = 5.04EE129 pKa = 5.12YY130 pKa = 10.01FGSYY134 pKa = 8.96IKK136 pKa = 9.34YY137 pKa = 9.28HH138 pKa = 6.31RR139 pKa = 11.84GIEE142 pKa = 4.09KK143 pKa = 10.4VLQQRR148 pKa = 11.84AKK150 pKa = 9.54PRR152 pKa = 11.84TWKK155 pKa = 10.63SRR157 pKa = 11.84VVVYY161 pKa = 8.91WGPTGTGKK169 pKa = 7.96TRR171 pKa = 11.84RR172 pKa = 11.84CFASSDD178 pKa = 3.78PPTWIYY184 pKa = 11.35SSDD187 pKa = 3.03GWFDD191 pKa = 3.99GYY193 pKa = 11.25QGDD196 pKa = 4.09EE197 pKa = 4.15TVLFDD202 pKa = 5.35DD203 pKa = 4.97FGGHH207 pKa = 5.76EE208 pKa = 4.34FRR210 pKa = 11.84LTHH213 pKa = 6.75LLKK216 pKa = 11.12LLDD219 pKa = 4.45RR220 pKa = 11.84YY221 pKa = 10.07PMRR224 pKa = 11.84VKK226 pKa = 10.8VKK228 pKa = 10.06GSFVQWCPKK237 pKa = 9.3TIYY240 pKa = 8.68ITSNIEE246 pKa = 3.47PAFWYY251 pKa = 9.37PNARR255 pKa = 11.84EE256 pKa = 4.05VHH258 pKa = 6.27RR259 pKa = 11.84EE260 pKa = 3.43ALMRR264 pKa = 11.84RR265 pKa = 11.84IDD267 pKa = 5.09DD268 pKa = 3.71IEE270 pKa = 4.07EE271 pKa = 4.17MADD274 pKa = 3.04EE275 pKa = 4.47WVPPVPVEE283 pKa = 4.31NEE285 pKa = 4.23VIDD288 pKa = 4.19LTDD291 pKa = 3.22EE292 pKa = 4.05
Molecular weight: 34.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5T695|S5T695_9CIRC Uncharacterized protein OS=Circoviridae 13 LDMD-2013 OX=1379717 PE=4 SV=1
MM1 pKa = 8.16PDD3 pKa = 3.15LDD5 pKa = 4.31QNFTYY10 pKa = 10.47SPPLPTFIGPCQVGPRR26 pKa = 11.84EE27 pKa = 3.97RR28 pKa = 11.84AVLGVSRR35 pKa = 11.84RR36 pKa = 11.84TCDD39 pKa = 2.9EE40 pKa = 3.82VAFRR44 pKa = 11.84DD45 pKa = 3.61YY46 pKa = 11.05FQIFRR51 pKa = 11.84AIYY54 pKa = 9.41IMLPKK59 pKa = 10.07RR60 pKa = 11.84IQQIKK65 pKa = 8.9PARR68 pKa = 11.84ANQGMLRR75 pKa = 11.84DD76 pKa = 3.65HH77 pKa = 6.53MRR79 pKa = 11.84AFMQTHH85 pKa = 6.95RR86 pKa = 11.84GSKK89 pKa = 10.03ARR91 pKa = 11.84IIAEE95 pKa = 4.05LASLGWHH102 pKa = 5.73ITPSFVKK109 pKa = 10.69GLGLSFGTWSATKK122 pKa = 10.48KK123 pKa = 10.17FIKK126 pKa = 10.3KK127 pKa = 9.84FEE129 pKa = 4.36KK130 pKa = 10.1PYY132 pKa = 10.34EE133 pKa = 4.14WDD135 pKa = 3.5DD136 pKa = 3.54APPRR140 pKa = 11.84VEE142 pKa = 4.6KK143 pKa = 10.69RR144 pKa = 11.84KK145 pKa = 9.08QSKK148 pKa = 10.07IKK150 pKa = 10.57FPSKK154 pKa = 8.76KK155 pKa = 9.35QKK157 pKa = 9.78RR158 pKa = 11.84VHH160 pKa = 6.32PTTQLLADD168 pKa = 3.79VAFGASKK175 pKa = 10.8KK176 pKa = 10.3SLSKK180 pKa = 9.79STYY183 pKa = 7.38TKK185 pKa = 10.29QMPYY189 pKa = 9.92RR190 pKa = 11.84RR191 pKa = 11.84KK192 pKa = 9.95GYY194 pKa = 10.18GKK196 pKa = 10.3SGMQKK201 pKa = 9.3RR202 pKa = 11.84RR203 pKa = 11.84RR204 pKa = 11.84YY205 pKa = 8.64KK206 pKa = 9.93KK207 pKa = 9.05KK208 pKa = 7.97RR209 pKa = 11.84QRR211 pKa = 11.84RR212 pKa = 11.84SRR214 pKa = 11.84SFGLTKK220 pKa = 10.3KK221 pKa = 10.14IKK223 pKa = 9.68RR224 pKa = 11.84QIRR227 pKa = 11.84NIAKK231 pKa = 9.98RR232 pKa = 11.84ISHH235 pKa = 6.62EE236 pKa = 4.25SLPQEE241 pKa = 3.75TRR243 pKa = 11.84EE244 pKa = 4.14IDD246 pKa = 3.07NHH248 pKa = 6.03QYY250 pKa = 10.92SSPVNTASYY259 pKa = 10.85NAIDD263 pKa = 4.32CLTRR267 pKa = 11.84DD268 pKa = 4.91DD269 pKa = 5.18VDD271 pKa = 3.45NLLNYY276 pKa = 7.8TYY278 pKa = 8.58PTIDD282 pKa = 4.05GGTIDD287 pKa = 4.24WKK289 pKa = 11.04DD290 pKa = 3.57LSSTNSDD297 pKa = 3.0CRR299 pKa = 11.84VYY301 pKa = 9.5FTKK304 pKa = 10.71GSYY307 pKa = 9.07LQLWIRR313 pKa = 11.84NNFDD317 pKa = 3.45YY318 pKa = 10.63PVTMKK323 pKa = 10.14FYY325 pKa = 10.08PYY327 pKa = 10.49KK328 pKa = 10.7CADD331 pKa = 3.59STNVDD336 pKa = 3.87AEE338 pKa = 4.41TEE340 pKa = 4.21MTNKK344 pKa = 9.63FQEE347 pKa = 4.31TQYY350 pKa = 11.57SVAAPPLILGNAKK363 pKa = 9.57QYY365 pKa = 11.39YY366 pKa = 9.21FINPSDD372 pKa = 3.77VKK374 pKa = 9.85MKK376 pKa = 10.06DD377 pKa = 2.93WKK379 pKa = 10.72RR380 pKa = 11.84HH381 pKa = 4.15GKK383 pKa = 9.41AQTKK387 pKa = 9.75IINPSEE393 pKa = 4.04EE394 pKa = 4.41CKK396 pKa = 10.93VSFPLTGYY404 pKa = 10.29YY405 pKa = 10.28DD406 pKa = 3.51YY407 pKa = 11.62RR408 pKa = 11.84NANDD412 pKa = 3.1QGTTYY417 pKa = 10.66IAGCSHH423 pKa = 6.75SLLIKK428 pKa = 10.29QYY430 pKa = 10.12GHH432 pKa = 7.34PSHH435 pKa = 6.74GKK437 pKa = 9.58QEE439 pKa = 3.84VDD441 pKa = 3.14AQGLNTGLSDD451 pKa = 3.5TKK453 pKa = 10.56VDD455 pKa = 4.02VISRR459 pKa = 11.84CNLKK463 pKa = 10.7YY464 pKa = 10.76KK465 pKa = 10.64LVTKK469 pKa = 10.47DD470 pKa = 3.34EE471 pKa = 4.47IGHH474 pKa = 6.2IKK476 pKa = 10.17HH477 pKa = 5.83GQNLDD482 pKa = 3.27RR483 pKa = 11.84YY484 pKa = 9.04GG485 pKa = 3.31
MM1 pKa = 8.16PDD3 pKa = 3.15LDD5 pKa = 4.31QNFTYY10 pKa = 10.47SPPLPTFIGPCQVGPRR26 pKa = 11.84EE27 pKa = 3.97RR28 pKa = 11.84AVLGVSRR35 pKa = 11.84RR36 pKa = 11.84TCDD39 pKa = 2.9EE40 pKa = 3.82VAFRR44 pKa = 11.84DD45 pKa = 3.61YY46 pKa = 11.05FQIFRR51 pKa = 11.84AIYY54 pKa = 9.41IMLPKK59 pKa = 10.07RR60 pKa = 11.84IQQIKK65 pKa = 8.9PARR68 pKa = 11.84ANQGMLRR75 pKa = 11.84DD76 pKa = 3.65HH77 pKa = 6.53MRR79 pKa = 11.84AFMQTHH85 pKa = 6.95RR86 pKa = 11.84GSKK89 pKa = 10.03ARR91 pKa = 11.84IIAEE95 pKa = 4.05LASLGWHH102 pKa = 5.73ITPSFVKK109 pKa = 10.69GLGLSFGTWSATKK122 pKa = 10.48KK123 pKa = 10.17FIKK126 pKa = 10.3KK127 pKa = 9.84FEE129 pKa = 4.36KK130 pKa = 10.1PYY132 pKa = 10.34EE133 pKa = 4.14WDD135 pKa = 3.5DD136 pKa = 3.54APPRR140 pKa = 11.84VEE142 pKa = 4.6KK143 pKa = 10.69RR144 pKa = 11.84KK145 pKa = 9.08QSKK148 pKa = 10.07IKK150 pKa = 10.57FPSKK154 pKa = 8.76KK155 pKa = 9.35QKK157 pKa = 9.78RR158 pKa = 11.84VHH160 pKa = 6.32PTTQLLADD168 pKa = 3.79VAFGASKK175 pKa = 10.8KK176 pKa = 10.3SLSKK180 pKa = 9.79STYY183 pKa = 7.38TKK185 pKa = 10.29QMPYY189 pKa = 9.92RR190 pKa = 11.84RR191 pKa = 11.84KK192 pKa = 9.95GYY194 pKa = 10.18GKK196 pKa = 10.3SGMQKK201 pKa = 9.3RR202 pKa = 11.84RR203 pKa = 11.84RR204 pKa = 11.84YY205 pKa = 8.64KK206 pKa = 9.93KK207 pKa = 9.05KK208 pKa = 7.97RR209 pKa = 11.84QRR211 pKa = 11.84RR212 pKa = 11.84SRR214 pKa = 11.84SFGLTKK220 pKa = 10.3KK221 pKa = 10.14IKK223 pKa = 9.68RR224 pKa = 11.84QIRR227 pKa = 11.84NIAKK231 pKa = 9.98RR232 pKa = 11.84ISHH235 pKa = 6.62EE236 pKa = 4.25SLPQEE241 pKa = 3.75TRR243 pKa = 11.84EE244 pKa = 4.14IDD246 pKa = 3.07NHH248 pKa = 6.03QYY250 pKa = 10.92SSPVNTASYY259 pKa = 10.85NAIDD263 pKa = 4.32CLTRR267 pKa = 11.84DD268 pKa = 4.91DD269 pKa = 5.18VDD271 pKa = 3.45NLLNYY276 pKa = 7.8TYY278 pKa = 8.58PTIDD282 pKa = 4.05GGTIDD287 pKa = 4.24WKK289 pKa = 11.04DD290 pKa = 3.57LSSTNSDD297 pKa = 3.0CRR299 pKa = 11.84VYY301 pKa = 9.5FTKK304 pKa = 10.71GSYY307 pKa = 9.07LQLWIRR313 pKa = 11.84NNFDD317 pKa = 3.45YY318 pKa = 10.63PVTMKK323 pKa = 10.14FYY325 pKa = 10.08PYY327 pKa = 10.49KK328 pKa = 10.7CADD331 pKa = 3.59STNVDD336 pKa = 3.87AEE338 pKa = 4.41TEE340 pKa = 4.21MTNKK344 pKa = 9.63FQEE347 pKa = 4.31TQYY350 pKa = 11.57SVAAPPLILGNAKK363 pKa = 9.57QYY365 pKa = 11.39YY366 pKa = 9.21FINPSDD372 pKa = 3.77VKK374 pKa = 9.85MKK376 pKa = 10.06DD377 pKa = 2.93WKK379 pKa = 10.72RR380 pKa = 11.84HH381 pKa = 4.15GKK383 pKa = 9.41AQTKK387 pKa = 9.75IINPSEE393 pKa = 4.04EE394 pKa = 4.41CKK396 pKa = 10.93VSFPLTGYY404 pKa = 10.29YY405 pKa = 10.28DD406 pKa = 3.51YY407 pKa = 11.62RR408 pKa = 11.84NANDD412 pKa = 3.1QGTTYY417 pKa = 10.66IAGCSHH423 pKa = 6.75SLLIKK428 pKa = 10.29QYY430 pKa = 10.12GHH432 pKa = 7.34PSHH435 pKa = 6.74GKK437 pKa = 9.58QEE439 pKa = 3.84VDD441 pKa = 3.14AQGLNTGLSDD451 pKa = 3.5TKK453 pKa = 10.56VDD455 pKa = 4.02VISRR459 pKa = 11.84CNLKK463 pKa = 10.7YY464 pKa = 10.76KK465 pKa = 10.64LVTKK469 pKa = 10.47DD470 pKa = 3.34EE471 pKa = 4.47IGHH474 pKa = 6.2IKK476 pKa = 10.17HH477 pKa = 5.83GQNLDD482 pKa = 3.27RR483 pKa = 11.84YY484 pKa = 9.04GG485 pKa = 3.31
Molecular weight: 55.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
777 |
292 |
485 |
388.5 |
45.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.663 ± 0.248 | 1.673 ± 0.019 |
5.92 ± 0.049 | 5.92 ± 2.151 |
4.247 ± 0.271 | 6.306 ± 0.268 |
2.703 ± 0.188 | 5.663 ± 0.26 |
8.366 ± 1.426 | 5.792 ± 0.323 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.931 ± 0.108 | 3.604 ± 0.596 |
5.148 ± 0.175 | 4.505 ± 0.703 |
7.593 ± 0.309 | 6.435 ± 0.641 |
6.049 ± 0.451 | 5.277 ± 0.946 |
1.802 ± 0.463 | 5.405 ± 0.133 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
