
Hubei leech virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.82
Get precalculated fractions of proteins
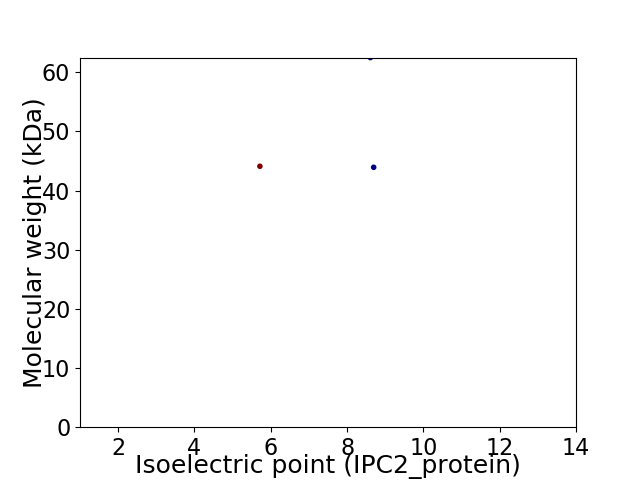
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
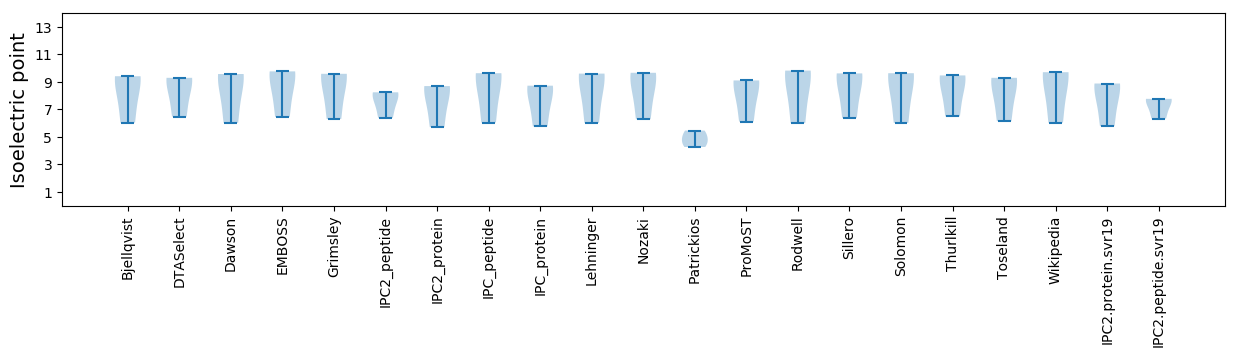
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KP21|A0A1L3KP21_9VIRU Putative capsid protein OS=Hubei leech virus 1 OX=1922899 PE=4 SV=1
MM1 pKa = 8.07DD2 pKa = 6.15LGNSPGWPWRR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 4.14VRR16 pKa = 11.84TKK18 pKa = 10.39QQAIDD23 pKa = 3.79AFPEE27 pKa = 3.96VLTYY31 pKa = 9.96WDD33 pKa = 3.52APGPVVWHH41 pKa = 6.26GFGKK45 pKa = 10.87LEE47 pKa = 4.02ILPSEE52 pKa = 4.24KK53 pKa = 10.15ADD55 pKa = 3.81RR56 pKa = 11.84KK57 pKa = 10.33CRR59 pKa = 11.84LITGAPVDD67 pKa = 4.23LALLGARR74 pKa = 11.84LYY76 pKa = 11.28EE77 pKa = 4.32DD78 pKa = 3.54FNDD81 pKa = 4.41RR82 pKa = 11.84FTTLHH87 pKa = 6.31TEE89 pKa = 3.85THH91 pKa = 6.11SALGMSKK98 pKa = 8.94FHH100 pKa = 7.03RR101 pKa = 11.84GWATLAAKK109 pKa = 9.65MDD111 pKa = 4.08GPVTEE116 pKa = 5.57ADD118 pKa = 3.19AKK120 pKa = 10.63QWDD123 pKa = 3.58SSMTIKK129 pKa = 10.78LLEE132 pKa = 3.93AVYY135 pKa = 10.22RR136 pKa = 11.84IRR138 pKa = 11.84WACLRR143 pKa = 11.84SEE145 pKa = 4.57DD146 pKa = 3.71QTAGNWARR154 pKa = 11.84HH155 pKa = 4.13QAYY158 pKa = 10.05LRR160 pKa = 11.84EE161 pKa = 4.37LTHH164 pKa = 5.88STIHH168 pKa = 6.75LPDD171 pKa = 4.0GSMWEE176 pKa = 4.15AIGGNKK182 pKa = 9.6SGSVNTAHH190 pKa = 7.62DD191 pKa = 3.72NTIGHH196 pKa = 7.31IIVCLYY202 pKa = 10.31AWLRR206 pKa = 11.84LGRR209 pKa = 11.84CSKK212 pKa = 10.41EE213 pKa = 4.19RR214 pKa = 11.84FWEE217 pKa = 4.47YY218 pKa = 10.77PFALYY223 pKa = 10.81GDD225 pKa = 4.67DD226 pKa = 4.45FLAGKK231 pKa = 10.29LEE233 pKa = 3.7EE234 pKa = 4.59RR235 pKa = 11.84FFDD238 pKa = 4.71FYY240 pKa = 11.28RR241 pKa = 11.84EE242 pKa = 3.83TGIRR246 pKa = 11.84LPRR249 pKa = 11.84EE250 pKa = 3.61NVVQHH255 pKa = 6.48DD256 pKa = 4.02QLEE259 pKa = 4.71GASFLSQRR267 pKa = 11.84FKK269 pKa = 11.08KK270 pKa = 10.68VDD272 pKa = 3.86GQWVAEE278 pKa = 4.02PDD280 pKa = 3.41GAKK283 pKa = 10.59ALFSACTSDD292 pKa = 4.47LKK294 pKa = 11.17PNLRR298 pKa = 11.84LAFEE302 pKa = 4.57RR303 pKa = 11.84VRR305 pKa = 11.84SLWLDD310 pKa = 3.72AYY312 pKa = 8.09WTEE315 pKa = 4.31AEE317 pKa = 4.08EE318 pKa = 4.32TLRR321 pKa = 11.84RR322 pKa = 11.84ACNSLADD329 pKa = 3.92EE330 pKa = 4.66LGEE333 pKa = 4.0PRR335 pKa = 11.84VSRR338 pKa = 11.84LLAKK342 pKa = 10.2RR343 pKa = 11.84IWNCYY348 pKa = 7.86EE349 pKa = 3.62IEE351 pKa = 4.61GGSGVSASPQNGEE364 pKa = 4.33HH365 pKa = 6.22NFITAADD372 pKa = 3.47HH373 pKa = 7.07CEE375 pKa = 3.91RR376 pKa = 11.84EE377 pKa = 4.63SVGEE381 pKa = 3.89NLPPPANLL389 pKa = 3.64
MM1 pKa = 8.07DD2 pKa = 6.15LGNSPGWPWRR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 4.14VRR16 pKa = 11.84TKK18 pKa = 10.39QQAIDD23 pKa = 3.79AFPEE27 pKa = 3.96VLTYY31 pKa = 9.96WDD33 pKa = 3.52APGPVVWHH41 pKa = 6.26GFGKK45 pKa = 10.87LEE47 pKa = 4.02ILPSEE52 pKa = 4.24KK53 pKa = 10.15ADD55 pKa = 3.81RR56 pKa = 11.84KK57 pKa = 10.33CRR59 pKa = 11.84LITGAPVDD67 pKa = 4.23LALLGARR74 pKa = 11.84LYY76 pKa = 11.28EE77 pKa = 4.32DD78 pKa = 3.54FNDD81 pKa = 4.41RR82 pKa = 11.84FTTLHH87 pKa = 6.31TEE89 pKa = 3.85THH91 pKa = 6.11SALGMSKK98 pKa = 8.94FHH100 pKa = 7.03RR101 pKa = 11.84GWATLAAKK109 pKa = 9.65MDD111 pKa = 4.08GPVTEE116 pKa = 5.57ADD118 pKa = 3.19AKK120 pKa = 10.63QWDD123 pKa = 3.58SSMTIKK129 pKa = 10.78LLEE132 pKa = 3.93AVYY135 pKa = 10.22RR136 pKa = 11.84IRR138 pKa = 11.84WACLRR143 pKa = 11.84SEE145 pKa = 4.57DD146 pKa = 3.71QTAGNWARR154 pKa = 11.84HH155 pKa = 4.13QAYY158 pKa = 10.05LRR160 pKa = 11.84EE161 pKa = 4.37LTHH164 pKa = 5.88STIHH168 pKa = 6.75LPDD171 pKa = 4.0GSMWEE176 pKa = 4.15AIGGNKK182 pKa = 9.6SGSVNTAHH190 pKa = 7.62DD191 pKa = 3.72NTIGHH196 pKa = 7.31IIVCLYY202 pKa = 10.31AWLRR206 pKa = 11.84LGRR209 pKa = 11.84CSKK212 pKa = 10.41EE213 pKa = 4.19RR214 pKa = 11.84FWEE217 pKa = 4.47YY218 pKa = 10.77PFALYY223 pKa = 10.81GDD225 pKa = 4.67DD226 pKa = 4.45FLAGKK231 pKa = 10.29LEE233 pKa = 3.7EE234 pKa = 4.59RR235 pKa = 11.84FFDD238 pKa = 4.71FYY240 pKa = 11.28RR241 pKa = 11.84EE242 pKa = 3.83TGIRR246 pKa = 11.84LPRR249 pKa = 11.84EE250 pKa = 3.61NVVQHH255 pKa = 6.48DD256 pKa = 4.02QLEE259 pKa = 4.71GASFLSQRR267 pKa = 11.84FKK269 pKa = 11.08KK270 pKa = 10.68VDD272 pKa = 3.86GQWVAEE278 pKa = 4.02PDD280 pKa = 3.41GAKK283 pKa = 10.59ALFSACTSDD292 pKa = 4.47LKK294 pKa = 11.17PNLRR298 pKa = 11.84LAFEE302 pKa = 4.57RR303 pKa = 11.84VRR305 pKa = 11.84SLWLDD310 pKa = 3.72AYY312 pKa = 8.09WTEE315 pKa = 4.31AEE317 pKa = 4.08EE318 pKa = 4.32TLRR321 pKa = 11.84RR322 pKa = 11.84ACNSLADD329 pKa = 3.92EE330 pKa = 4.66LGEE333 pKa = 4.0PRR335 pKa = 11.84VSRR338 pKa = 11.84LLAKK342 pKa = 10.2RR343 pKa = 11.84IWNCYY348 pKa = 7.86EE349 pKa = 3.62IEE351 pKa = 4.61GGSGVSASPQNGEE364 pKa = 4.33HH365 pKa = 6.22NFITAADD372 pKa = 3.47HH373 pKa = 7.07CEE375 pKa = 3.91RR376 pKa = 11.84EE377 pKa = 4.63SVGEE381 pKa = 3.89NLPPPANLL389 pKa = 3.64
Molecular weight: 44.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KNW4|A0A1L3KNW4_9VIRU RNA-dependent RNA polymerase OS=Hubei leech virus 1 OX=1922899 PE=4 SV=1
MM1 pKa = 7.55TYY3 pKa = 10.49WNIKK7 pKa = 10.04VIMTILLLGVAAVAVIAGVLFVCFKK32 pKa = 10.9VRR34 pKa = 11.84VSTQQAASLTSTFIGFCVFLWIVRR58 pKa = 11.84ALVRR62 pKa = 11.84RR63 pKa = 11.84YY64 pKa = 10.12GPNKK68 pKa = 8.3TVRR71 pKa = 11.84EE72 pKa = 3.96LVMEE76 pKa = 4.48RR77 pKa = 11.84FKK79 pKa = 11.2KK80 pKa = 10.2KK81 pKa = 9.71RR82 pKa = 11.84SKK84 pKa = 10.15AAKK87 pKa = 9.57GPSGTMAGDD96 pKa = 3.62QYY98 pKa = 11.34QVNLIPGVTDD108 pKa = 4.38LLSEE112 pKa = 4.2LWDD115 pKa = 4.28DD116 pKa = 4.21VDD118 pKa = 4.67DD119 pKa = 4.39TFTLAVPLFAMGGVAWGRR137 pKa = 11.84DD138 pKa = 3.3LLFIQTAIRR147 pKa = 11.84KK148 pKa = 8.12FVATLPGWLDD158 pKa = 3.6LPLGDD163 pKa = 4.68LRR165 pKa = 11.84GRR167 pKa = 11.84GIAFVQRR174 pKa = 11.84MHH176 pKa = 7.12DD177 pKa = 3.46QGGDD181 pKa = 3.56PAQGPQPQRR190 pKa = 11.84PWRR193 pKa = 11.84EE194 pKa = 3.55RR195 pKa = 11.84LRR197 pKa = 11.84DD198 pKa = 3.53RR199 pKa = 11.84AIQLYY204 pKa = 9.59EE205 pKa = 4.49AGTHH209 pKa = 6.5PIARR213 pKa = 11.84AWDD216 pKa = 3.42EE217 pKa = 4.12AQRR220 pKa = 11.84LMDD223 pKa = 4.15GPNYY227 pKa = 10.24RR228 pKa = 11.84ATLKK232 pKa = 10.6KK233 pKa = 10.11IAVTGMTACALFLAYY248 pKa = 10.19RR249 pKa = 11.84SWSGRR254 pKa = 11.84KK255 pKa = 8.9NGKK258 pKa = 8.89KK259 pKa = 10.05EE260 pKa = 3.78RR261 pKa = 11.84DD262 pKa = 3.89EE263 pKa = 5.13PEE265 pKa = 3.44MDD267 pKa = 4.04QIVPEE272 pKa = 4.93DD273 pKa = 3.83KK274 pKa = 10.69TPSPLMLRR282 pKa = 11.84EE283 pKa = 4.0VFMSGSTVLQNVPRR297 pKa = 11.84AVQGVRR303 pKa = 11.84LLRR306 pKa = 11.84LPEE309 pKa = 4.38NGSTRR314 pKa = 11.84QSYY317 pKa = 6.9MTCFKK322 pKa = 10.51VGRR325 pKa = 11.84FFVTCSHH332 pKa = 6.01VEE334 pKa = 3.93FGEE337 pKa = 4.41GEE339 pKa = 3.9EE340 pKa = 4.11AAIALEE346 pKa = 4.12AVPLDD351 pKa = 4.94SNPTWLRR358 pKa = 11.84ARR360 pKa = 11.84VLKK363 pKa = 9.85VFPRR367 pKa = 11.84RR368 pKa = 11.84TCEE371 pKa = 3.65LADD374 pKa = 4.91DD375 pKa = 4.26IVVLEE380 pKa = 5.16CPAQLATVRR389 pKa = 11.84SIPSRR394 pKa = 11.84VAKK397 pKa = 9.54EE398 pKa = 3.58NPIAGFVAGFVDD410 pKa = 5.38GVHH413 pKa = 7.07SIPAMSPGAIDD424 pKa = 3.21TEE426 pKa = 4.67GTHH429 pKa = 6.12WCTTAPGASGSAVCDD444 pKa = 3.52HH445 pKa = 7.14AGVAIGVHH453 pKa = 5.2YY454 pKa = 10.75AGSHH458 pKa = 5.78KK459 pKa = 10.66LNSFMPFDD467 pKa = 4.16RR468 pKa = 11.84EE469 pKa = 3.77VLDD472 pKa = 4.18FVKK475 pKa = 10.71GGNSEE480 pKa = 3.91PFEE483 pKa = 4.53RR484 pKa = 11.84SVKK487 pKa = 8.96RR488 pKa = 11.84TRR490 pKa = 11.84ASSQPNGGRR499 pKa = 11.84TPGLSPLQEE508 pKa = 4.03PTLEE512 pKa = 4.15ISDD515 pKa = 3.92TSGLDD520 pKa = 3.36PLPNLSGPKK529 pKa = 9.62SSSSTATSPPTDD541 pKa = 3.46AAPISRR547 pKa = 11.84SRR549 pKa = 11.84RR550 pKa = 11.84RR551 pKa = 11.84RR552 pKa = 11.84LARR555 pKa = 11.84RR556 pKa = 11.84LKK558 pKa = 10.44KK559 pKa = 10.9LSDD562 pKa = 3.51TCSEE566 pKa = 4.21SSANAGG572 pKa = 3.34
MM1 pKa = 7.55TYY3 pKa = 10.49WNIKK7 pKa = 10.04VIMTILLLGVAAVAVIAGVLFVCFKK32 pKa = 10.9VRR34 pKa = 11.84VSTQQAASLTSTFIGFCVFLWIVRR58 pKa = 11.84ALVRR62 pKa = 11.84RR63 pKa = 11.84YY64 pKa = 10.12GPNKK68 pKa = 8.3TVRR71 pKa = 11.84EE72 pKa = 3.96LVMEE76 pKa = 4.48RR77 pKa = 11.84FKK79 pKa = 11.2KK80 pKa = 10.2KK81 pKa = 9.71RR82 pKa = 11.84SKK84 pKa = 10.15AAKK87 pKa = 9.57GPSGTMAGDD96 pKa = 3.62QYY98 pKa = 11.34QVNLIPGVTDD108 pKa = 4.38LLSEE112 pKa = 4.2LWDD115 pKa = 4.28DD116 pKa = 4.21VDD118 pKa = 4.67DD119 pKa = 4.39TFTLAVPLFAMGGVAWGRR137 pKa = 11.84DD138 pKa = 3.3LLFIQTAIRR147 pKa = 11.84KK148 pKa = 8.12FVATLPGWLDD158 pKa = 3.6LPLGDD163 pKa = 4.68LRR165 pKa = 11.84GRR167 pKa = 11.84GIAFVQRR174 pKa = 11.84MHH176 pKa = 7.12DD177 pKa = 3.46QGGDD181 pKa = 3.56PAQGPQPQRR190 pKa = 11.84PWRR193 pKa = 11.84EE194 pKa = 3.55RR195 pKa = 11.84LRR197 pKa = 11.84DD198 pKa = 3.53RR199 pKa = 11.84AIQLYY204 pKa = 9.59EE205 pKa = 4.49AGTHH209 pKa = 6.5PIARR213 pKa = 11.84AWDD216 pKa = 3.42EE217 pKa = 4.12AQRR220 pKa = 11.84LMDD223 pKa = 4.15GPNYY227 pKa = 10.24RR228 pKa = 11.84ATLKK232 pKa = 10.6KK233 pKa = 10.11IAVTGMTACALFLAYY248 pKa = 10.19RR249 pKa = 11.84SWSGRR254 pKa = 11.84KK255 pKa = 8.9NGKK258 pKa = 8.89KK259 pKa = 10.05EE260 pKa = 3.78RR261 pKa = 11.84DD262 pKa = 3.89EE263 pKa = 5.13PEE265 pKa = 3.44MDD267 pKa = 4.04QIVPEE272 pKa = 4.93DD273 pKa = 3.83KK274 pKa = 10.69TPSPLMLRR282 pKa = 11.84EE283 pKa = 4.0VFMSGSTVLQNVPRR297 pKa = 11.84AVQGVRR303 pKa = 11.84LLRR306 pKa = 11.84LPEE309 pKa = 4.38NGSTRR314 pKa = 11.84QSYY317 pKa = 6.9MTCFKK322 pKa = 10.51VGRR325 pKa = 11.84FFVTCSHH332 pKa = 6.01VEE334 pKa = 3.93FGEE337 pKa = 4.41GEE339 pKa = 3.9EE340 pKa = 4.11AAIALEE346 pKa = 4.12AVPLDD351 pKa = 4.94SNPTWLRR358 pKa = 11.84ARR360 pKa = 11.84VLKK363 pKa = 9.85VFPRR367 pKa = 11.84RR368 pKa = 11.84TCEE371 pKa = 3.65LADD374 pKa = 4.91DD375 pKa = 4.26IVVLEE380 pKa = 5.16CPAQLATVRR389 pKa = 11.84SIPSRR394 pKa = 11.84VAKK397 pKa = 9.54EE398 pKa = 3.58NPIAGFVAGFVDD410 pKa = 5.38GVHH413 pKa = 7.07SIPAMSPGAIDD424 pKa = 3.21TEE426 pKa = 4.67GTHH429 pKa = 6.12WCTTAPGASGSAVCDD444 pKa = 3.52HH445 pKa = 7.14AGVAIGVHH453 pKa = 5.2YY454 pKa = 10.75AGSHH458 pKa = 5.78KK459 pKa = 10.66LNSFMPFDD467 pKa = 4.16RR468 pKa = 11.84EE469 pKa = 3.77VLDD472 pKa = 4.18FVKK475 pKa = 10.71GGNSEE480 pKa = 3.91PFEE483 pKa = 4.53RR484 pKa = 11.84SVKK487 pKa = 8.96RR488 pKa = 11.84TRR490 pKa = 11.84ASSQPNGGRR499 pKa = 11.84TPGLSPLQEE508 pKa = 4.03PTLEE512 pKa = 4.15ISDD515 pKa = 3.92TSGLDD520 pKa = 3.36PLPNLSGPKK529 pKa = 9.62SSSSTATSPPTDD541 pKa = 3.46AAPISRR547 pKa = 11.84SRR549 pKa = 11.84RR550 pKa = 11.84RR551 pKa = 11.84RR552 pKa = 11.84LARR555 pKa = 11.84RR556 pKa = 11.84LKK558 pKa = 10.44KK559 pKa = 10.9LSDD562 pKa = 3.51TCSEE566 pKa = 4.21SSANAGG572 pKa = 3.34
Molecular weight: 62.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1370 |
389 |
572 |
456.7 |
50.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.781 ± 0.216 | 1.533 ± 0.316 |
4.745 ± 0.63 | 5.401 ± 0.839 |
3.723 ± 0.41 | 8.321 ± 0.408 |
1.898 ± 0.521 | 3.65 ± 0.221 |
3.796 ± 0.342 | 8.978 ± 0.537 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.117 ± 0.3 | 2.774 ± 0.298 |
6.423 ± 0.668 | 3.869 ± 0.791 |
7.445 ± 0.551 | 7.518 ± 0.749 |
6.35 ± 0.535 | 7.007 ± 0.952 |
2.628 ± 0.519 | 2.044 ± 0.341 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
