
Sanxia permutotetra-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.72
Get precalculated fractions of proteins
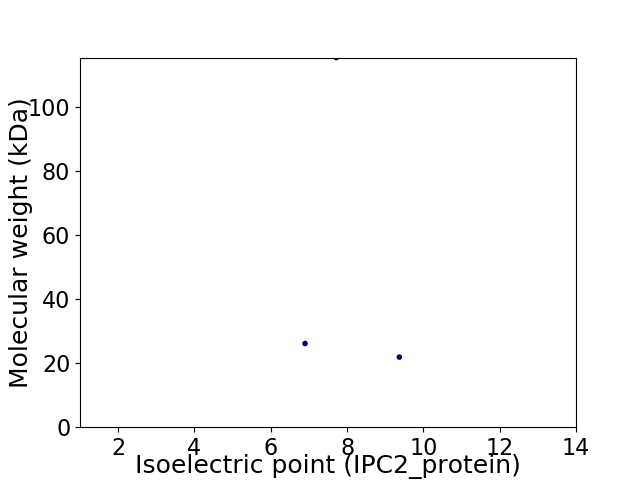
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHX9|A0A1L3KHX9_9VIRU Capsid protein OS=Sanxia permutotetra-like virus 1 OX=1923365 PE=3 SV=1
MM1 pKa = 7.05TANALDD7 pKa = 3.86GAAAKK12 pKa = 10.57SKK14 pKa = 10.85GPDD17 pKa = 3.99PIQTLHH23 pKa = 7.2LGDD26 pKa = 3.68NTLSLEE32 pKa = 4.26SRR34 pKa = 11.84NTPAFDD40 pKa = 4.14NISRR44 pKa = 11.84INLGLDD50 pKa = 2.99TFLTIRR56 pKa = 11.84TNFTLNAGDD65 pKa = 3.58ITSVVCSLKK74 pKa = 10.96SSDD77 pKa = 4.07GIPWNIPWTGVPVVDD92 pKa = 5.51KK93 pKa = 8.92FVCEE97 pKa = 4.15SGKK100 pKa = 10.4KK101 pKa = 9.78FYY103 pKa = 11.14SVTTNLLLRR112 pKa = 11.84CDD114 pKa = 3.96KK115 pKa = 10.93ARR117 pKa = 11.84SANFYY122 pKa = 10.46FEE124 pKa = 4.41WEE126 pKa = 4.04VDD128 pKa = 3.78FTTGFRR134 pKa = 11.84AGWNFVMQYY143 pKa = 9.19TLDD146 pKa = 5.07LIASPYY152 pKa = 10.54LNLSILPAIGYY163 pKa = 7.61GTQQYY168 pKa = 8.85QAILDD173 pKa = 4.11GLHH176 pKa = 6.72HH177 pKa = 7.11EE178 pKa = 4.5EE179 pKa = 4.94AQDD182 pKa = 3.68QAKK185 pKa = 8.55STQPVISNLYY195 pKa = 8.0PQLPTAPQVDD205 pKa = 3.7TRR207 pKa = 11.84MEE209 pKa = 4.55ADD211 pKa = 3.74LQQPPIKK218 pKa = 9.67TKK220 pKa = 10.9AKK222 pKa = 10.3LLTRR226 pKa = 11.84LFKK229 pKa = 10.83RR230 pKa = 11.84KK231 pKa = 9.2ISKK234 pKa = 9.57PP235 pKa = 3.42
MM1 pKa = 7.05TANALDD7 pKa = 3.86GAAAKK12 pKa = 10.57SKK14 pKa = 10.85GPDD17 pKa = 3.99PIQTLHH23 pKa = 7.2LGDD26 pKa = 3.68NTLSLEE32 pKa = 4.26SRR34 pKa = 11.84NTPAFDD40 pKa = 4.14NISRR44 pKa = 11.84INLGLDD50 pKa = 2.99TFLTIRR56 pKa = 11.84TNFTLNAGDD65 pKa = 3.58ITSVVCSLKK74 pKa = 10.96SSDD77 pKa = 4.07GIPWNIPWTGVPVVDD92 pKa = 5.51KK93 pKa = 8.92FVCEE97 pKa = 4.15SGKK100 pKa = 10.4KK101 pKa = 9.78FYY103 pKa = 11.14SVTTNLLLRR112 pKa = 11.84CDD114 pKa = 3.96KK115 pKa = 10.93ARR117 pKa = 11.84SANFYY122 pKa = 10.46FEE124 pKa = 4.41WEE126 pKa = 4.04VDD128 pKa = 3.78FTTGFRR134 pKa = 11.84AGWNFVMQYY143 pKa = 9.19TLDD146 pKa = 5.07LIASPYY152 pKa = 10.54LNLSILPAIGYY163 pKa = 7.61GTQQYY168 pKa = 8.85QAILDD173 pKa = 4.11GLHH176 pKa = 6.72HH177 pKa = 7.11EE178 pKa = 4.5EE179 pKa = 4.94AQDD182 pKa = 3.68QAKK185 pKa = 8.55STQPVISNLYY195 pKa = 8.0PQLPTAPQVDD205 pKa = 3.7TRR207 pKa = 11.84MEE209 pKa = 4.55ADD211 pKa = 3.74LQQPPIKK218 pKa = 9.67TKK220 pKa = 10.9AKK222 pKa = 10.3LLTRR226 pKa = 11.84LFKK229 pKa = 10.83RR230 pKa = 11.84KK231 pKa = 9.2ISKK234 pKa = 9.57PP235 pKa = 3.42
Molecular weight: 26.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHX9|A0A1L3KHX9_9VIRU Capsid protein OS=Sanxia permutotetra-like virus 1 OX=1923365 PE=3 SV=1
MM1 pKa = 7.86PNNKK5 pKa = 9.22NMNKK9 pKa = 10.07RR10 pKa = 11.84GQKK13 pKa = 9.39PQAKK17 pKa = 9.07KK18 pKa = 10.3QKK20 pKa = 10.17LPRR23 pKa = 11.84KK24 pKa = 7.25TVAGQPKK31 pKa = 9.94AVNLLMGVPVTGRR44 pKa = 11.84EE45 pKa = 3.81VTAILTASANKK56 pKa = 9.88TSVSYY61 pKa = 11.71NNTFNIASASPMLKK75 pKa = 9.86KK76 pKa = 10.39YY77 pKa = 10.14AAIYY81 pKa = 8.69EE82 pKa = 4.37SYY84 pKa = 10.24RR85 pKa = 11.84IKK87 pKa = 10.61SVSYY91 pKa = 10.37RR92 pKa = 11.84FIPDD96 pKa = 3.16EE97 pKa = 4.6SGLSSGNISLGIDD110 pKa = 3.36YY111 pKa = 10.94GKK113 pKa = 10.1MSGTLTRR120 pKa = 11.84EE121 pKa = 3.96QISRR125 pKa = 11.84LNPHH129 pKa = 6.05YY130 pKa = 10.5SGPIRR135 pKa = 11.84KK136 pKa = 7.79ITPWVTIHH144 pKa = 6.64PKK146 pKa = 9.97FVNKK150 pKa = 9.27DD151 pKa = 3.36TVRR154 pKa = 11.84YY155 pKa = 10.26SGDD158 pKa = 3.71DD159 pKa = 3.5SLLSAPFTLALVATCEE175 pKa = 4.28GKK177 pKa = 10.49SVDD180 pKa = 3.65RR181 pKa = 11.84VLGCVEE187 pKa = 3.6IQYY190 pKa = 9.03TLEE193 pKa = 4.02FQGILPP199 pKa = 4.34
MM1 pKa = 7.86PNNKK5 pKa = 9.22NMNKK9 pKa = 10.07RR10 pKa = 11.84GQKK13 pKa = 9.39PQAKK17 pKa = 9.07KK18 pKa = 10.3QKK20 pKa = 10.17LPRR23 pKa = 11.84KK24 pKa = 7.25TVAGQPKK31 pKa = 9.94AVNLLMGVPVTGRR44 pKa = 11.84EE45 pKa = 3.81VTAILTASANKK56 pKa = 9.88TSVSYY61 pKa = 11.71NNTFNIASASPMLKK75 pKa = 9.86KK76 pKa = 10.39YY77 pKa = 10.14AAIYY81 pKa = 8.69EE82 pKa = 4.37SYY84 pKa = 10.24RR85 pKa = 11.84IKK87 pKa = 10.61SVSYY91 pKa = 10.37RR92 pKa = 11.84FIPDD96 pKa = 3.16EE97 pKa = 4.6SGLSSGNISLGIDD110 pKa = 3.36YY111 pKa = 10.94GKK113 pKa = 10.1MSGTLTRR120 pKa = 11.84EE121 pKa = 3.96QISRR125 pKa = 11.84LNPHH129 pKa = 6.05YY130 pKa = 10.5SGPIRR135 pKa = 11.84KK136 pKa = 7.79ITPWVTIHH144 pKa = 6.64PKK146 pKa = 9.97FVNKK150 pKa = 9.27DD151 pKa = 3.36TVRR154 pKa = 11.84YY155 pKa = 10.26SGDD158 pKa = 3.71DD159 pKa = 3.5SLLSAPFTLALVATCEE175 pKa = 4.28GKK177 pKa = 10.49SVDD180 pKa = 3.65RR181 pKa = 11.84VLGCVEE187 pKa = 3.6IQYY190 pKa = 9.03TLEE193 pKa = 4.02FQGILPP199 pKa = 4.34
Molecular weight: 21.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1461 |
199 |
1027 |
487.0 |
54.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.666 ± 0.323 | 0.821 ± 0.176 |
5.818 ± 0.731 | 5.133 ± 0.991 |
3.217 ± 0.415 | 5.749 ± 0.339 |
1.574 ± 0.223 | 5.065 ± 0.606 |
7.255 ± 0.406 | 9.514 ± 0.477 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.669 ± 0.468 | 3.696 ± 1.072 |
5.886 ± 0.294 | 4.997 ± 0.406 |
5.133 ± 0.432 | 6.639 ± 0.854 |
7.666 ± 0.36 | 6.845 ± 0.669 |
1.506 ± 0.262 | 3.149 ± 0.369 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
