
Arthrobacter sp. SW1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Arthrobacter; unclassified Arthrobacter
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
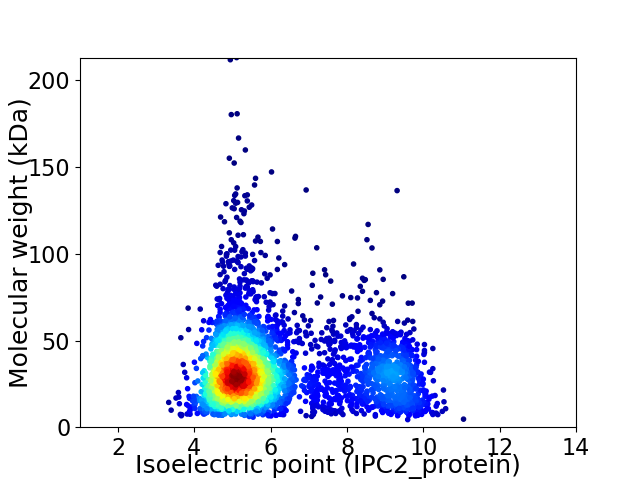
Virtual 2D-PAGE plot for 3661 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
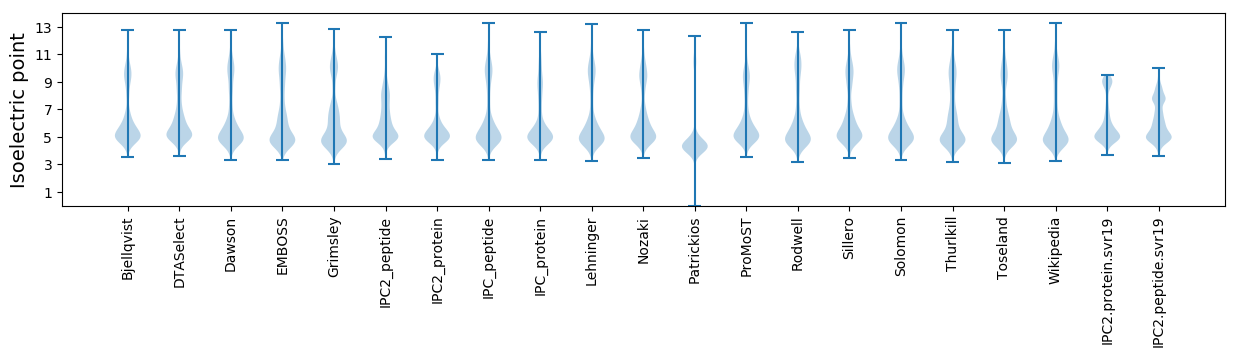
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E8FNB8|A0A1E8FNB8_9MICC Transcriptional regulator OS=Arthrobacter sp. SW1 OX=1920889 GN=BIU82_10275 PE=4 SV=1
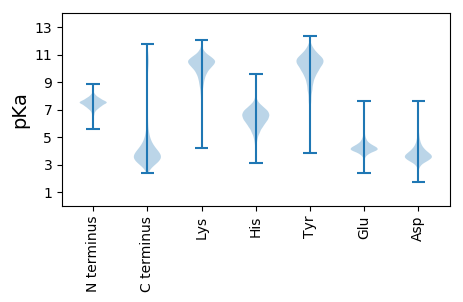
MM1 pKa = 7.2VFLAALSTLALSTFGLTGASADD23 pKa = 3.71PDD25 pKa = 4.01GQHH28 pKa = 6.87GSDD31 pKa = 3.56EE32 pKa = 4.14GHH34 pKa = 7.42LIGTGATGKK43 pKa = 9.57IQLLDD48 pKa = 3.71VVDD51 pKa = 4.11VTDD54 pKa = 4.19TPDD57 pKa = 4.39LIADD61 pKa = 3.8VTVSPDD67 pKa = 2.82GNTAFLANWGEE78 pKa = 4.02PDD80 pKa = 3.55CAGPEE85 pKa = 4.08TGGQNSPDD93 pKa = 3.18AGAYY97 pKa = 10.18VIDD100 pKa = 3.7ITNIEE105 pKa = 4.36TDD107 pKa = 3.6PEE109 pKa = 4.31SAKK112 pKa = 10.92LIGFIPSHH120 pKa = 6.27QDD122 pKa = 2.95TRR124 pKa = 11.84PGEE127 pKa = 4.14GMQVVNVTTAQFSGDD142 pKa = 3.2ILVMNNEE149 pKa = 4.18ACGKK153 pKa = 7.78NAKK156 pKa = 10.17GGVSLWDD163 pKa = 3.32VTDD166 pKa = 3.71PANPKK171 pKa = 9.96KK172 pKa = 10.6LSEE175 pKa = 4.26HH176 pKa = 6.22FGDD179 pKa = 4.36RR180 pKa = 11.84EE181 pKa = 4.03PGTRR185 pKa = 11.84DD186 pKa = 2.99ANEE189 pKa = 3.72IHH191 pKa = 7.12SAFAWDD197 pKa = 4.07AGNNAYY203 pKa = 10.72LVMTDD208 pKa = 3.75NEE210 pKa = 4.25QFPDD214 pKa = 3.64VDD216 pKa = 4.4ILDD219 pKa = 3.53ITNPHH224 pKa = 6.78RR225 pKa = 11.84PRR227 pKa = 11.84LIAEE231 pKa = 4.03YY232 pKa = 10.22NLNDD236 pKa = 3.83FAVDD240 pKa = 3.74QPEE243 pKa = 4.22LGLTEE248 pKa = 4.92SFLHH252 pKa = 6.65DD253 pKa = 3.67MVVKK257 pKa = 10.54EE258 pKa = 3.6IDD260 pKa = 3.19GRR262 pKa = 11.84FIMLLSYY269 pKa = 10.21WDD271 pKa = 3.57GGYY274 pKa = 10.86VLLDD278 pKa = 3.27VTDD281 pKa = 3.92PANAVFIGDD290 pKa = 3.62TDD292 pKa = 4.29FAAVDD297 pKa = 3.67PEE299 pKa = 4.37LFEE302 pKa = 4.53SLGVALTPEE311 pKa = 4.76GNGHH315 pKa = 5.0QAEE318 pKa = 4.36FTKK321 pKa = 11.06DD322 pKa = 2.91NAFFIGTDD330 pKa = 3.29EE331 pKa = 5.03DD332 pKa = 3.87FAPYY336 pKa = 8.79RR337 pKa = 11.84TDD339 pKa = 3.07ALRR342 pKa = 11.84ITSGAFQGEE351 pKa = 4.68YY352 pKa = 10.45EE353 pKa = 4.41SVIVPGGQPPAVLEE367 pKa = 4.4DD368 pKa = 3.84LTLSGPVVYY377 pKa = 10.22GGYY380 pKa = 10.05GCPASAPVPSPEE392 pKa = 4.37DD393 pKa = 3.33VPGYY397 pKa = 9.76VASLQPGDD405 pKa = 3.64EE406 pKa = 4.63TILVLQRR413 pKa = 11.84GPSGDD418 pKa = 3.53PSATEE423 pKa = 3.87EE424 pKa = 4.07ACFPGEE430 pKa = 4.17KK431 pKa = 9.73AHH433 pKa = 6.42EE434 pKa = 4.21AALAGWDD441 pKa = 3.03AVLFVQRR448 pKa = 11.84HH449 pKa = 4.77LPAGEE454 pKa = 4.41EE455 pKa = 3.97DD456 pKa = 3.73TAFCGSGAFVDD467 pKa = 5.16SIVAVCTTHH476 pKa = 6.86EE477 pKa = 4.26AFHH480 pKa = 6.76KK481 pKa = 10.56LFNLEE486 pKa = 4.38PLDD489 pKa = 4.12PAQWSYY495 pKa = 11.63PEE497 pKa = 4.13NLAIGAIGDD506 pKa = 4.43RR507 pKa = 11.84IRR509 pKa = 11.84VGSIFDD515 pKa = 3.07GWGYY519 pKa = 10.71VHH521 pKa = 7.34LFDD524 pKa = 4.95TATLQEE530 pKa = 4.75LDD532 pKa = 3.47TYY534 pKa = 10.83AIPEE538 pKa = 3.94AHH540 pKa = 7.03DD541 pKa = 3.2PAFAFGYY548 pKa = 10.52GDD550 pKa = 3.97LSVHH554 pKa = 5.78EE555 pKa = 4.57VAVDD559 pKa = 3.68PQDD562 pKa = 3.59PSLAYY567 pKa = 10.33LSYY570 pKa = 11.08YY571 pKa = 10.66SGGLRR576 pKa = 11.84AIQIQCGGVPYY587 pKa = 10.76DD588 pKa = 4.55ADD590 pKa = 3.93NPPADD595 pKa = 4.07KK596 pKa = 10.85GSCEE600 pKa = 3.93LVEE603 pKa = 4.28VGSYY607 pKa = 10.9LDD609 pKa = 3.49PAGNDD614 pKa = 3.07FWGVEE619 pKa = 4.16TFIGDD624 pKa = 3.83DD625 pKa = 3.15GRR627 pKa = 11.84TYY629 pKa = 11.0ILGSDD634 pKa = 3.72RR635 pKa = 11.84DD636 pKa = 3.64SGLWIFTDD644 pKa = 3.86PP645 pKa = 4.44
MM1 pKa = 7.2VFLAALSTLALSTFGLTGASADD23 pKa = 3.71PDD25 pKa = 4.01GQHH28 pKa = 6.87GSDD31 pKa = 3.56EE32 pKa = 4.14GHH34 pKa = 7.42LIGTGATGKK43 pKa = 9.57IQLLDD48 pKa = 3.71VVDD51 pKa = 4.11VTDD54 pKa = 4.19TPDD57 pKa = 4.39LIADD61 pKa = 3.8VTVSPDD67 pKa = 2.82GNTAFLANWGEE78 pKa = 4.02PDD80 pKa = 3.55CAGPEE85 pKa = 4.08TGGQNSPDD93 pKa = 3.18AGAYY97 pKa = 10.18VIDD100 pKa = 3.7ITNIEE105 pKa = 4.36TDD107 pKa = 3.6PEE109 pKa = 4.31SAKK112 pKa = 10.92LIGFIPSHH120 pKa = 6.27QDD122 pKa = 2.95TRR124 pKa = 11.84PGEE127 pKa = 4.14GMQVVNVTTAQFSGDD142 pKa = 3.2ILVMNNEE149 pKa = 4.18ACGKK153 pKa = 7.78NAKK156 pKa = 10.17GGVSLWDD163 pKa = 3.32VTDD166 pKa = 3.71PANPKK171 pKa = 9.96KK172 pKa = 10.6LSEE175 pKa = 4.26HH176 pKa = 6.22FGDD179 pKa = 4.36RR180 pKa = 11.84EE181 pKa = 4.03PGTRR185 pKa = 11.84DD186 pKa = 2.99ANEE189 pKa = 3.72IHH191 pKa = 7.12SAFAWDD197 pKa = 4.07AGNNAYY203 pKa = 10.72LVMTDD208 pKa = 3.75NEE210 pKa = 4.25QFPDD214 pKa = 3.64VDD216 pKa = 4.4ILDD219 pKa = 3.53ITNPHH224 pKa = 6.78RR225 pKa = 11.84PRR227 pKa = 11.84LIAEE231 pKa = 4.03YY232 pKa = 10.22NLNDD236 pKa = 3.83FAVDD240 pKa = 3.74QPEE243 pKa = 4.22LGLTEE248 pKa = 4.92SFLHH252 pKa = 6.65DD253 pKa = 3.67MVVKK257 pKa = 10.54EE258 pKa = 3.6IDD260 pKa = 3.19GRR262 pKa = 11.84FIMLLSYY269 pKa = 10.21WDD271 pKa = 3.57GGYY274 pKa = 10.86VLLDD278 pKa = 3.27VTDD281 pKa = 3.92PANAVFIGDD290 pKa = 3.62TDD292 pKa = 4.29FAAVDD297 pKa = 3.67PEE299 pKa = 4.37LFEE302 pKa = 4.53SLGVALTPEE311 pKa = 4.76GNGHH315 pKa = 5.0QAEE318 pKa = 4.36FTKK321 pKa = 11.06DD322 pKa = 2.91NAFFIGTDD330 pKa = 3.29EE331 pKa = 5.03DD332 pKa = 3.87FAPYY336 pKa = 8.79RR337 pKa = 11.84TDD339 pKa = 3.07ALRR342 pKa = 11.84ITSGAFQGEE351 pKa = 4.68YY352 pKa = 10.45EE353 pKa = 4.41SVIVPGGQPPAVLEE367 pKa = 4.4DD368 pKa = 3.84LTLSGPVVYY377 pKa = 10.22GGYY380 pKa = 10.05GCPASAPVPSPEE392 pKa = 4.37DD393 pKa = 3.33VPGYY397 pKa = 9.76VASLQPGDD405 pKa = 3.64EE406 pKa = 4.63TILVLQRR413 pKa = 11.84GPSGDD418 pKa = 3.53PSATEE423 pKa = 3.87EE424 pKa = 4.07ACFPGEE430 pKa = 4.17KK431 pKa = 9.73AHH433 pKa = 6.42EE434 pKa = 4.21AALAGWDD441 pKa = 3.03AVLFVQRR448 pKa = 11.84HH449 pKa = 4.77LPAGEE454 pKa = 4.41EE455 pKa = 3.97DD456 pKa = 3.73TAFCGSGAFVDD467 pKa = 5.16SIVAVCTTHH476 pKa = 6.86EE477 pKa = 4.26AFHH480 pKa = 6.76KK481 pKa = 10.56LFNLEE486 pKa = 4.38PLDD489 pKa = 4.12PAQWSYY495 pKa = 11.63PEE497 pKa = 4.13NLAIGAIGDD506 pKa = 4.43RR507 pKa = 11.84IRR509 pKa = 11.84VGSIFDD515 pKa = 3.07GWGYY519 pKa = 10.71VHH521 pKa = 7.34LFDD524 pKa = 4.95TATLQEE530 pKa = 4.75LDD532 pKa = 3.47TYY534 pKa = 10.83AIPEE538 pKa = 3.94AHH540 pKa = 7.03DD541 pKa = 3.2PAFAFGYY548 pKa = 10.52GDD550 pKa = 3.97LSVHH554 pKa = 5.78EE555 pKa = 4.57VAVDD559 pKa = 3.68PQDD562 pKa = 3.59PSLAYY567 pKa = 10.33LSYY570 pKa = 11.08YY571 pKa = 10.66SGGLRR576 pKa = 11.84AIQIQCGGVPYY587 pKa = 10.76DD588 pKa = 4.55ADD590 pKa = 3.93NPPADD595 pKa = 4.07KK596 pKa = 10.85GSCEE600 pKa = 3.93LVEE603 pKa = 4.28VGSYY607 pKa = 10.9LDD609 pKa = 3.49PAGNDD614 pKa = 3.07FWGVEE619 pKa = 4.16TFIGDD624 pKa = 3.83DD625 pKa = 3.15GRR627 pKa = 11.84TYY629 pKa = 11.0ILGSDD634 pKa = 3.72RR635 pKa = 11.84DD636 pKa = 3.64SGLWIFTDD644 pKa = 3.86PP645 pKa = 4.44
Molecular weight: 68.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E8FVW3|A0A1E8FVW3_9MICC AB hydrolase-1 domain-containing protein OS=Arthrobacter sp. SW1 OX=1920889 GN=BIU82_02175 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 10.31VRR4 pKa = 11.84NSLRR8 pKa = 11.84ALKK11 pKa = 9.61KK12 pKa = 9.72IPGAQVVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GKK24 pKa = 8.59TFVINKK30 pKa = 8.84KK31 pKa = 9.15NPRR34 pKa = 11.84FKK36 pKa = 10.85ARR38 pKa = 11.84QGG40 pKa = 3.28
MM1 pKa = 7.61KK2 pKa = 10.31VRR4 pKa = 11.84NSLRR8 pKa = 11.84ALKK11 pKa = 9.61KK12 pKa = 9.72IPGAQVVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GKK24 pKa = 8.59TFVINKK30 pKa = 8.84KK31 pKa = 9.15NPRR34 pKa = 11.84FKK36 pKa = 10.85ARR38 pKa = 11.84QGG40 pKa = 3.28
Molecular weight: 4.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1202471 |
37 |
2047 |
328.5 |
35.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.352 ± 0.059 | 0.625 ± 0.009 |
5.444 ± 0.036 | 5.87 ± 0.046 |
3.334 ± 0.03 | 9.295 ± 0.038 |
2.009 ± 0.019 | 4.279 ± 0.033 |
2.965 ± 0.033 | 10.381 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.978 ± 0.018 | 2.428 ± 0.024 |
5.438 ± 0.033 | 2.96 ± 0.022 |
6.409 ± 0.042 | 5.846 ± 0.025 |
5.55 ± 0.033 | 8.331 ± 0.037 |
1.415 ± 0.018 | 2.089 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
