
Candidatus Izimaplasma sp. HR2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Tenericutes incertae sedis; Candidatus Izimaplasma; unclassified Candidatus Izimaplasma
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
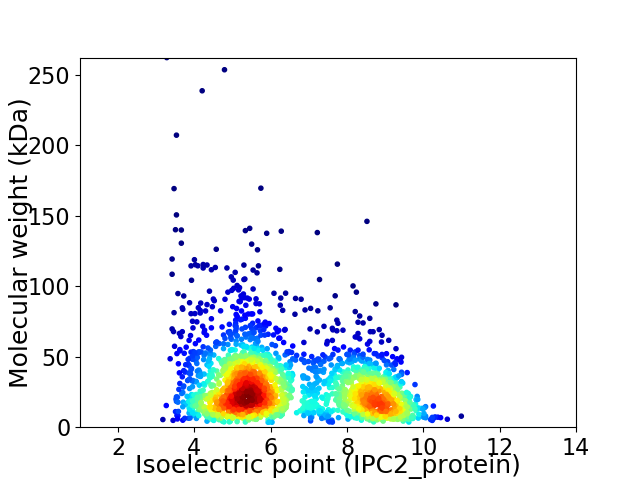
Virtual 2D-PAGE plot for 2222 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
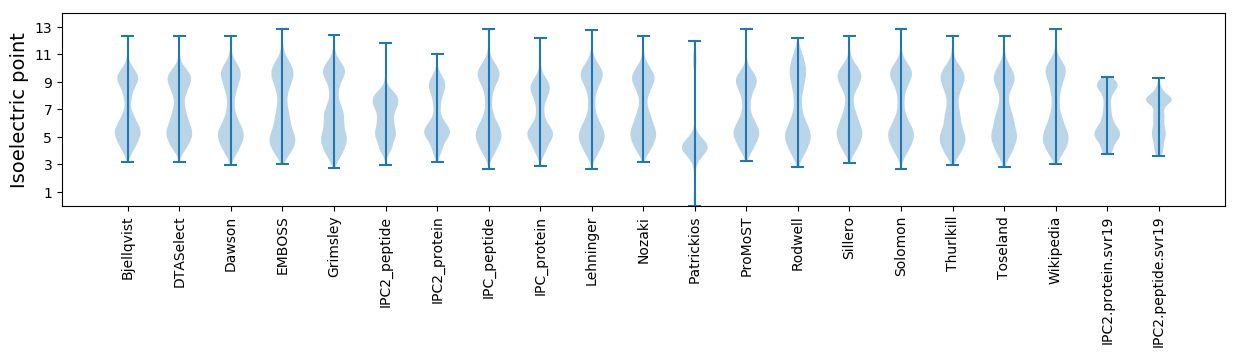
Protein with the lowest isoelectric point:
>tr|A0A094ILE8|A0A094ILE8_9BACT Uncharacterized protein OS=Candidatus Izimaplasma sp. HR2 OX=1541960 GN=KQ78_01144 PE=4 SV=1
MM1 pKa = 7.37IKK3 pKa = 10.17KK4 pKa = 8.23ITMALTVFILALTLTGCNGNSLSDD28 pKa = 3.58TEE30 pKa = 4.26QVLEE34 pKa = 4.32IKK36 pKa = 10.83NSISFDD42 pKa = 3.54LLEE45 pKa = 5.26IEE47 pKa = 4.52NNITIPSSDD56 pKa = 3.4IEE58 pKa = 4.65GVTLTWVSSKK68 pKa = 10.39PQYY71 pKa = 11.02LDD73 pKa = 2.71IDD75 pKa = 4.04GTVTIPTASMGDD87 pKa = 3.56QNVDD91 pKa = 3.23LSVTIEE97 pKa = 4.07FGTAVTTKK105 pKa = 9.43VFHH108 pKa = 6.14FTVIAPVVVIDD119 pKa = 3.69TALNTDD125 pKa = 3.26QTDD128 pKa = 3.58ALAMDD133 pKa = 4.6FTYY136 pKa = 11.14EE137 pKa = 4.0DD138 pKa = 3.49TDD140 pKa = 4.6FIASGIGEE148 pKa = 4.4VTLVSCIDD156 pKa = 3.44GDD158 pKa = 4.0TAIFTEE164 pKa = 5.15GGANFTVRR172 pKa = 11.84FLGVDD177 pKa = 3.73TPEE180 pKa = 3.55STYY183 pKa = 11.06RR184 pKa = 11.84FDD186 pKa = 3.55PWGKK190 pKa = 8.13AASMFTCDD198 pKa = 4.02KK199 pKa = 9.44LTNATTIVLEE209 pKa = 4.1YY210 pKa = 11.03DD211 pKa = 3.34PATTRR216 pKa = 11.84TEE218 pKa = 4.01GNGRR222 pKa = 11.84YY223 pKa = 9.34LSWIWYY229 pKa = 9.22DD230 pKa = 3.49GRR232 pKa = 11.84LLNLEE237 pKa = 5.0LIEE240 pKa = 4.15QAFSGSKK247 pKa = 10.18GVAGLKK253 pKa = 10.11YY254 pKa = 10.58EE255 pKa = 4.07NLFYY259 pKa = 9.04TTEE262 pKa = 3.93FTVQDD267 pKa = 3.33KK268 pKa = 10.86DD269 pKa = 3.5RR270 pKa = 11.84RR271 pKa = 11.84IWGEE275 pKa = 3.77DD276 pKa = 3.45DD277 pKa = 3.72PDD279 pKa = 3.57YY280 pKa = 11.16DD281 pKa = 3.94YY282 pKa = 11.87SLDD285 pKa = 3.58GVQITIEE292 pKa = 4.03EE293 pKa = 4.53LVTNQEE299 pKa = 4.48LYY301 pKa = 10.88VGTKK305 pKa = 9.32VVITGVVSRR314 pKa = 11.84NLEE317 pKa = 3.83GHH319 pKa = 7.25PYY321 pKa = 10.15LQVGDD326 pKa = 3.94YY327 pKa = 10.95AIYY330 pKa = 10.28LYY332 pKa = 10.08IGYY335 pKa = 7.99TYY337 pKa = 7.54TTKK340 pKa = 10.56LVEE343 pKa = 4.46GNEE346 pKa = 4.09VTISGLTPTYY356 pKa = 10.63HH357 pKa = 7.53PDD359 pKa = 3.26AEE361 pKa = 4.57TGSLQLTGFTRR372 pKa = 11.84EE373 pKa = 4.23HH374 pKa = 5.69ITVISVGNVVEE385 pKa = 4.14PEE387 pKa = 3.9IIEE390 pKa = 3.96VSEE393 pKa = 3.68IDD395 pKa = 3.93IYY397 pKa = 10.87TLSKK401 pKa = 10.79LITIEE406 pKa = 4.03NLTVTDD412 pKa = 4.49IYY414 pKa = 11.73VNDD417 pKa = 3.41ITGDD421 pKa = 3.64FTVTAEE427 pKa = 4.35DD428 pKa = 3.99SSDD431 pKa = 3.21NTISIRR437 pKa = 11.84INSGTDD443 pKa = 3.06ALLLPEE449 pKa = 5.35LFTVGTIFDD458 pKa = 3.67VTGPLGRR465 pKa = 11.84YY466 pKa = 7.01EE467 pKa = 4.57GNYY470 pKa = 9.4QVLLIYY476 pKa = 10.73YY477 pKa = 10.12DD478 pKa = 4.3DD479 pKa = 4.27VDD481 pKa = 4.95FKK483 pKa = 11.75
MM1 pKa = 7.37IKK3 pKa = 10.17KK4 pKa = 8.23ITMALTVFILALTLTGCNGNSLSDD28 pKa = 3.58TEE30 pKa = 4.26QVLEE34 pKa = 4.32IKK36 pKa = 10.83NSISFDD42 pKa = 3.54LLEE45 pKa = 5.26IEE47 pKa = 4.52NNITIPSSDD56 pKa = 3.4IEE58 pKa = 4.65GVTLTWVSSKK68 pKa = 10.39PQYY71 pKa = 11.02LDD73 pKa = 2.71IDD75 pKa = 4.04GTVTIPTASMGDD87 pKa = 3.56QNVDD91 pKa = 3.23LSVTIEE97 pKa = 4.07FGTAVTTKK105 pKa = 9.43VFHH108 pKa = 6.14FTVIAPVVVIDD119 pKa = 3.69TALNTDD125 pKa = 3.26QTDD128 pKa = 3.58ALAMDD133 pKa = 4.6FTYY136 pKa = 11.14EE137 pKa = 4.0DD138 pKa = 3.49TDD140 pKa = 4.6FIASGIGEE148 pKa = 4.4VTLVSCIDD156 pKa = 3.44GDD158 pKa = 4.0TAIFTEE164 pKa = 5.15GGANFTVRR172 pKa = 11.84FLGVDD177 pKa = 3.73TPEE180 pKa = 3.55STYY183 pKa = 11.06RR184 pKa = 11.84FDD186 pKa = 3.55PWGKK190 pKa = 8.13AASMFTCDD198 pKa = 4.02KK199 pKa = 9.44LTNATTIVLEE209 pKa = 4.1YY210 pKa = 11.03DD211 pKa = 3.34PATTRR216 pKa = 11.84TEE218 pKa = 4.01GNGRR222 pKa = 11.84YY223 pKa = 9.34LSWIWYY229 pKa = 9.22DD230 pKa = 3.49GRR232 pKa = 11.84LLNLEE237 pKa = 5.0LIEE240 pKa = 4.15QAFSGSKK247 pKa = 10.18GVAGLKK253 pKa = 10.11YY254 pKa = 10.58EE255 pKa = 4.07NLFYY259 pKa = 9.04TTEE262 pKa = 3.93FTVQDD267 pKa = 3.33KK268 pKa = 10.86DD269 pKa = 3.5RR270 pKa = 11.84RR271 pKa = 11.84IWGEE275 pKa = 3.77DD276 pKa = 3.45DD277 pKa = 3.72PDD279 pKa = 3.57YY280 pKa = 11.16DD281 pKa = 3.94YY282 pKa = 11.87SLDD285 pKa = 3.58GVQITIEE292 pKa = 4.03EE293 pKa = 4.53LVTNQEE299 pKa = 4.48LYY301 pKa = 10.88VGTKK305 pKa = 9.32VVITGVVSRR314 pKa = 11.84NLEE317 pKa = 3.83GHH319 pKa = 7.25PYY321 pKa = 10.15LQVGDD326 pKa = 3.94YY327 pKa = 10.95AIYY330 pKa = 10.28LYY332 pKa = 10.08IGYY335 pKa = 7.99TYY337 pKa = 7.54TTKK340 pKa = 10.56LVEE343 pKa = 4.46GNEE346 pKa = 4.09VTISGLTPTYY356 pKa = 10.63HH357 pKa = 7.53PDD359 pKa = 3.26AEE361 pKa = 4.57TGSLQLTGFTRR372 pKa = 11.84EE373 pKa = 4.23HH374 pKa = 5.69ITVISVGNVVEE385 pKa = 4.14PEE387 pKa = 3.9IIEE390 pKa = 3.96VSEE393 pKa = 3.68IDD395 pKa = 3.93IYY397 pKa = 10.87TLSKK401 pKa = 10.79LITIEE406 pKa = 4.03NLTVTDD412 pKa = 4.49IYY414 pKa = 11.73VNDD417 pKa = 3.41ITGDD421 pKa = 3.64FTVTAEE427 pKa = 4.35DD428 pKa = 3.99SSDD431 pKa = 3.21NTISIRR437 pKa = 11.84INSGTDD443 pKa = 3.06ALLLPEE449 pKa = 5.35LFTVGTIFDD458 pKa = 3.67VTGPLGRR465 pKa = 11.84YY466 pKa = 7.01EE467 pKa = 4.57GNYY470 pKa = 9.4QVLLIYY476 pKa = 10.73YY477 pKa = 10.12DD478 pKa = 4.3DD479 pKa = 4.27VDD481 pKa = 4.95FKK483 pKa = 11.75
Molecular weight: 53.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A094J4E0|A0A094J4E0_9BACT Putative hydrolase OS=Candidatus Izimaplasma sp. HR2 OX=1541960 GN=KQ78_00426 PE=4 SV=1
MM1 pKa = 7.6AKK3 pKa = 10.21KK4 pKa = 10.68AMIVKK9 pKa = 8.18QQRR12 pKa = 11.84GSKK15 pKa = 10.13FSTRR19 pKa = 11.84EE20 pKa = 3.41YY21 pKa = 10.13TRR23 pKa = 11.84CEE25 pKa = 3.81RR26 pKa = 11.84CGRR29 pKa = 11.84PHH31 pKa = 7.24AVYY34 pKa = 10.5KK35 pKa = 10.51KK36 pKa = 10.43FKK38 pKa = 10.08LCRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.94LAYY49 pKa = 10.67LGQIPGVRR57 pKa = 11.84KK58 pKa = 10.13ASWW61 pKa = 2.78
MM1 pKa = 7.6AKK3 pKa = 10.21KK4 pKa = 10.68AMIVKK9 pKa = 8.18QQRR12 pKa = 11.84GSKK15 pKa = 10.13FSTRR19 pKa = 11.84EE20 pKa = 3.41YY21 pKa = 10.13TRR23 pKa = 11.84CEE25 pKa = 3.81RR26 pKa = 11.84CGRR29 pKa = 11.84PHH31 pKa = 7.24AVYY34 pKa = 10.5KK35 pKa = 10.51KK36 pKa = 10.43FKK38 pKa = 10.08LCRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.94LAYY49 pKa = 10.67LGQIPGVRR57 pKa = 11.84KK58 pKa = 10.13ASWW61 pKa = 2.78
Molecular weight: 7.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
623788 |
30 |
2441 |
280.7 |
31.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.039 ± 0.06 | 0.837 ± 0.025 |
6.148 ± 0.048 | 7.118 ± 0.054 |
4.89 ± 0.044 | 6.107 ± 0.059 |
1.636 ± 0.024 | 9.841 ± 0.057 |
8.077 ± 0.089 | 9.399 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.548 ± 0.027 | 5.956 ± 0.052 |
2.689 ± 0.031 | 2.312 ± 0.03 |
3.414 ± 0.043 | 6.331 ± 0.048 |
5.506 ± 0.061 | 6.731 ± 0.051 |
0.693 ± 0.018 | 4.716 ± 0.045 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
