
Vitrella brassicaformis (strain CCMP3155)
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Colpodellida; Vitrellaceae; Vitrella; Vitrella brassicaformis
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
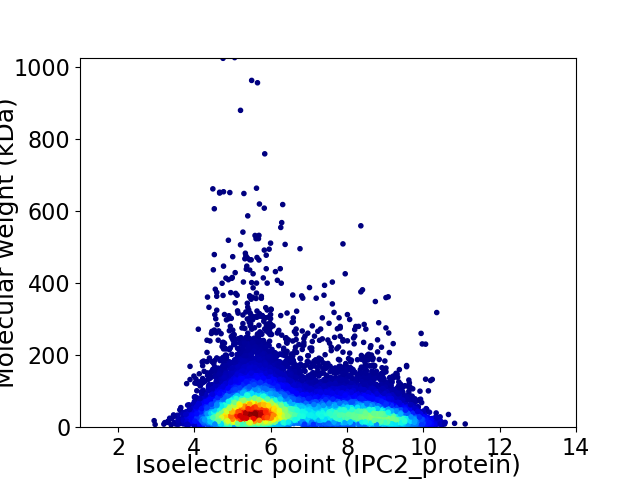
Virtual 2D-PAGE plot for 23030 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G4EEP3|A0A0G4EEP3_VITBC Uncharacterized protein OS=Vitrella brassicaformis (strain CCMP3155) OX=1169540 GN=Vbra_11403 PE=4 SV=1
MM1 pKa = 7.77AARR4 pKa = 11.84PQPGVLYY11 pKa = 10.44RR12 pKa = 11.84VDD14 pKa = 4.55DD15 pKa = 4.39DD16 pKa = 4.3PGVRR20 pKa = 11.84RR21 pKa = 11.84GGDD24 pKa = 3.38LEE26 pKa = 4.47RR27 pKa = 11.84YY28 pKa = 9.08GYY30 pKa = 10.48QRR32 pKa = 11.84TALEE36 pKa = 3.8DD37 pKa = 3.32HH38 pKa = 6.53VRR40 pKa = 11.84FGSQQRR46 pKa = 11.84STWISTSKK54 pKa = 10.73SPSAALYY61 pKa = 10.37YY62 pKa = 10.55GLRR65 pKa = 11.84DD66 pKa = 3.31EE67 pKa = 5.22NIRR70 pKa = 11.84DD71 pKa = 3.4KK72 pKa = 10.55TLLVIRR78 pKa = 11.84PDD80 pKa = 3.78DD81 pKa = 3.93GNRR84 pKa = 11.84GHH86 pKa = 7.12DD87 pKa = 3.45VSGRR91 pKa = 11.84LPDD94 pKa = 3.53QRR96 pKa = 11.84AHH98 pKa = 6.14NFADD102 pKa = 3.47ASEE105 pKa = 4.13EE106 pKa = 4.42VVFDD110 pKa = 3.43SCIPAQCIVDD120 pKa = 4.05RR121 pKa = 11.84VPLSEE126 pKa = 5.83LLDD129 pKa = 3.85EE130 pKa = 5.14LDD132 pKa = 3.79IDD134 pKa = 4.09EE135 pKa = 4.95GDD137 pKa = 3.99ARR139 pKa = 11.84DD140 pKa = 3.62MAQSLGVRR148 pKa = 11.84DD149 pKa = 4.03FRR151 pKa = 11.84EE152 pKa = 4.06AMEE155 pKa = 5.15GYY157 pKa = 8.76YY158 pKa = 9.66QVEE161 pKa = 3.83GRR163 pKa = 11.84MDD165 pKa = 3.51EE166 pKa = 4.59LYY168 pKa = 9.19ATGRR172 pKa = 11.84PVHH175 pKa = 6.76SSTAGDD181 pKa = 3.38GGYY184 pKa = 10.35DD185 pKa = 3.45EE186 pKa = 5.96PSYY189 pKa = 11.62DD190 pKa = 3.43WADD193 pKa = 3.76PPDD196 pKa = 4.11GAEE199 pKa = 3.94SPVYY203 pKa = 9.89RR204 pKa = 11.84VGGFTVLDD212 pKa = 4.42DD213 pKa = 4.85EE214 pKa = 6.11PDD216 pKa = 3.32DD217 pKa = 3.99TYY219 pKa = 11.89EE220 pKa = 3.83NDD222 pKa = 3.56YY223 pKa = 10.74EE224 pKa = 4.61NEE226 pKa = 4.09VAHH229 pKa = 6.72TGGLWMVDD237 pKa = 3.69DD238 pKa = 5.27DD239 pKa = 5.34QDD241 pKa = 3.69PAANGPYY248 pKa = 9.73DD249 pKa = 4.8DD250 pKa = 4.36EE251 pKa = 5.25PSYY254 pKa = 11.06VGHH257 pKa = 6.99IGGFYY262 pKa = 10.28IVDD265 pKa = 3.83EE266 pKa = 4.55QPAEE270 pKa = 3.92ALSAGRR276 pKa = 11.84SYY278 pKa = 11.65GDD280 pKa = 3.37QDD282 pKa = 4.21SEE284 pKa = 4.45DD285 pKa = 4.23DD286 pKa = 3.7EE287 pKa = 5.91GEE289 pKa = 4.0EE290 pKa = 4.57SEE292 pKa = 4.8VNSGSHH298 pKa = 6.87DD299 pKa = 3.93DD300 pKa = 4.53GYY302 pKa = 11.8GSYY305 pKa = 10.47EE306 pKa = 3.98VVEE309 pKa = 4.18EE310 pKa = 5.12HH311 pKa = 7.01VDD313 pKa = 3.58QSDD316 pKa = 3.45AAAAAADD323 pKa = 3.92GAEE326 pKa = 4.37EE327 pKa = 4.65GGGDD331 pKa = 3.79PEE333 pKa = 4.2YY334 pKa = 10.87DD335 pKa = 3.33YY336 pKa = 11.88GGNDD340 pKa = 3.12GDD342 pKa = 4.83AYY344 pKa = 10.7GAADD348 pKa = 4.32DD349 pKa = 4.98GGYY352 pKa = 10.12SDD354 pKa = 4.73GYY356 pKa = 10.47GWGSEE361 pKa = 4.38GGDD364 pKa = 3.82SDD366 pKa = 3.58WSAA369 pKa = 3.46
MM1 pKa = 7.77AARR4 pKa = 11.84PQPGVLYY11 pKa = 10.44RR12 pKa = 11.84VDD14 pKa = 4.55DD15 pKa = 4.39DD16 pKa = 4.3PGVRR20 pKa = 11.84RR21 pKa = 11.84GGDD24 pKa = 3.38LEE26 pKa = 4.47RR27 pKa = 11.84YY28 pKa = 9.08GYY30 pKa = 10.48QRR32 pKa = 11.84TALEE36 pKa = 3.8DD37 pKa = 3.32HH38 pKa = 6.53VRR40 pKa = 11.84FGSQQRR46 pKa = 11.84STWISTSKK54 pKa = 10.73SPSAALYY61 pKa = 10.37YY62 pKa = 10.55GLRR65 pKa = 11.84DD66 pKa = 3.31EE67 pKa = 5.22NIRR70 pKa = 11.84DD71 pKa = 3.4KK72 pKa = 10.55TLLVIRR78 pKa = 11.84PDD80 pKa = 3.78DD81 pKa = 3.93GNRR84 pKa = 11.84GHH86 pKa = 7.12DD87 pKa = 3.45VSGRR91 pKa = 11.84LPDD94 pKa = 3.53QRR96 pKa = 11.84AHH98 pKa = 6.14NFADD102 pKa = 3.47ASEE105 pKa = 4.13EE106 pKa = 4.42VVFDD110 pKa = 3.43SCIPAQCIVDD120 pKa = 4.05RR121 pKa = 11.84VPLSEE126 pKa = 5.83LLDD129 pKa = 3.85EE130 pKa = 5.14LDD132 pKa = 3.79IDD134 pKa = 4.09EE135 pKa = 4.95GDD137 pKa = 3.99ARR139 pKa = 11.84DD140 pKa = 3.62MAQSLGVRR148 pKa = 11.84DD149 pKa = 4.03FRR151 pKa = 11.84EE152 pKa = 4.06AMEE155 pKa = 5.15GYY157 pKa = 8.76YY158 pKa = 9.66QVEE161 pKa = 3.83GRR163 pKa = 11.84MDD165 pKa = 3.51EE166 pKa = 4.59LYY168 pKa = 9.19ATGRR172 pKa = 11.84PVHH175 pKa = 6.76SSTAGDD181 pKa = 3.38GGYY184 pKa = 10.35DD185 pKa = 3.45EE186 pKa = 5.96PSYY189 pKa = 11.62DD190 pKa = 3.43WADD193 pKa = 3.76PPDD196 pKa = 4.11GAEE199 pKa = 3.94SPVYY203 pKa = 9.89RR204 pKa = 11.84VGGFTVLDD212 pKa = 4.42DD213 pKa = 4.85EE214 pKa = 6.11PDD216 pKa = 3.32DD217 pKa = 3.99TYY219 pKa = 11.89EE220 pKa = 3.83NDD222 pKa = 3.56YY223 pKa = 10.74EE224 pKa = 4.61NEE226 pKa = 4.09VAHH229 pKa = 6.72TGGLWMVDD237 pKa = 3.69DD238 pKa = 5.27DD239 pKa = 5.34QDD241 pKa = 3.69PAANGPYY248 pKa = 9.73DD249 pKa = 4.8DD250 pKa = 4.36EE251 pKa = 5.25PSYY254 pKa = 11.06VGHH257 pKa = 6.99IGGFYY262 pKa = 10.28IVDD265 pKa = 3.83EE266 pKa = 4.55QPAEE270 pKa = 3.92ALSAGRR276 pKa = 11.84SYY278 pKa = 11.65GDD280 pKa = 3.37QDD282 pKa = 4.21SEE284 pKa = 4.45DD285 pKa = 4.23DD286 pKa = 3.7EE287 pKa = 5.91GEE289 pKa = 4.0EE290 pKa = 4.57SEE292 pKa = 4.8VNSGSHH298 pKa = 6.87DD299 pKa = 3.93DD300 pKa = 4.53GYY302 pKa = 11.8GSYY305 pKa = 10.47EE306 pKa = 3.98VVEE309 pKa = 4.18EE310 pKa = 5.12HH311 pKa = 7.01VDD313 pKa = 3.58QSDD316 pKa = 3.45AAAAAADD323 pKa = 3.92GAEE326 pKa = 4.37EE327 pKa = 4.65GGGDD331 pKa = 3.79PEE333 pKa = 4.2YY334 pKa = 10.87DD335 pKa = 3.33YY336 pKa = 11.88GGNDD340 pKa = 3.12GDD342 pKa = 4.83AYY344 pKa = 10.7GAADD348 pKa = 4.32DD349 pKa = 4.98GGYY352 pKa = 10.12SDD354 pKa = 4.73GYY356 pKa = 10.47GWGSEE361 pKa = 4.38GGDD364 pKa = 3.82SDD366 pKa = 3.58WSAA369 pKa = 3.46
Molecular weight: 40.04 kDa
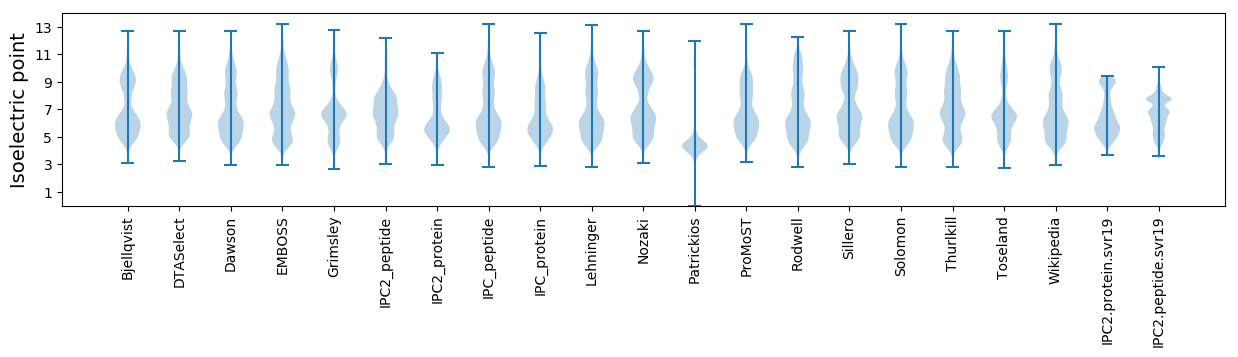
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G4EN05|A0A0G4EN05_VITBC Uncharacterized protein OS=Vitrella brassicaformis (strain CCMP3155) OX=1169540 GN=Vbra_22906 PE=4 SV=1
MM1 pKa = 7.06ATSRR5 pKa = 11.84YY6 pKa = 10.19ARR8 pKa = 11.84MLQLWWLTLPHH19 pKa = 5.58RR20 pKa = 11.84QKK22 pKa = 10.48RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84MGSSRR30 pKa = 11.84PLPQFCRR37 pKa = 11.84VVPPPLPRR45 pKa = 11.84HH46 pKa = 5.86HH47 pKa = 6.95SRR49 pKa = 11.84PASRR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 5.23RR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84SRR60 pKa = 11.84QTRR63 pKa = 11.84QNLSVRR69 pKa = 11.84RR70 pKa = 11.84WII72 pKa = 4.1
MM1 pKa = 7.06ATSRR5 pKa = 11.84YY6 pKa = 10.19ARR8 pKa = 11.84MLQLWWLTLPHH19 pKa = 5.58RR20 pKa = 11.84QKK22 pKa = 10.48RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84MGSSRR30 pKa = 11.84PLPQFCRR37 pKa = 11.84VVPPPLPRR45 pKa = 11.84HH46 pKa = 5.86HH47 pKa = 6.95SRR49 pKa = 11.84PASRR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 5.23RR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84SRR60 pKa = 11.84QTRR63 pKa = 11.84QNLSVRR69 pKa = 11.84RR70 pKa = 11.84WII72 pKa = 4.1
Molecular weight: 8.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11959242 |
19 |
9609 |
519.3 |
57.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.602 ± 0.019 | 1.701 ± 0.009 |
5.942 ± 0.012 | 6.353 ± 0.015 |
3.172 ± 0.011 | 7.649 ± 0.017 |
2.806 ± 0.008 | 3.683 ± 0.011 |
4.169 ± 0.016 | 8.856 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.313 ± 0.006 | 2.552 ± 0.009 |
6.776 ± 0.02 | 4.362 ± 0.013 |
7.073 ± 0.017 | 7.455 ± 0.015 |
5.502 ± 0.013 | 6.459 ± 0.013 |
1.359 ± 0.006 | 2.183 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
