
Phenylobacterium soli
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Phenylobacterium
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
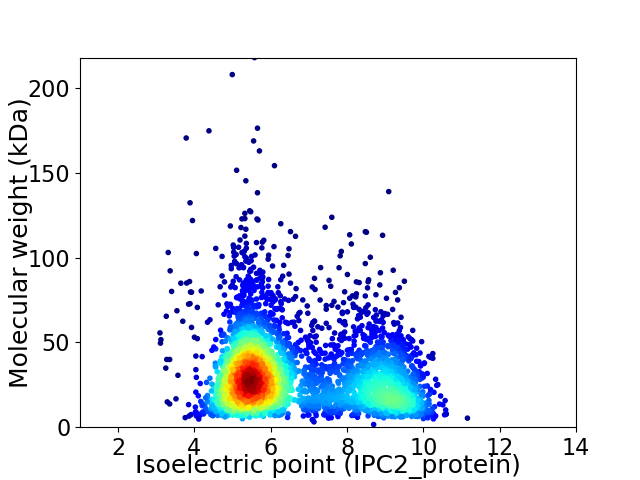
Virtual 2D-PAGE plot for 3905 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A328AMB1|A0A328AMB1_9CAUL Peptidase_S8 domain-containing protein OS=Phenylobacterium soli OX=2170551 GN=DJ017_11645 PE=4 SV=1
MM1 pKa = 7.86PSGPSGSLLVTPGGPITDD19 pKa = 3.4STYY22 pKa = 8.35TARR25 pKa = 11.84SFRR28 pKa = 11.84LTNTSTDD35 pKa = 3.44GQQLNTVTIDD45 pKa = 2.91ISGSVLGGMIYY56 pKa = 10.59DD57 pKa = 3.86PTGQAGDD64 pKa = 3.51STARR68 pKa = 11.84PFTIDD73 pKa = 3.09SQTGTFSLVNATLSNGSDD91 pKa = 2.86ATGYY95 pKa = 6.68TTLTLQLSGFDD106 pKa = 3.89PGEE109 pKa = 4.06EE110 pKa = 4.15LDD112 pKa = 4.53FSIDD116 pKa = 3.17VDD118 pKa = 4.01PNSIRR123 pKa = 11.84GTTSPGPGEE132 pKa = 4.39SGSVAGVEE140 pKa = 4.23LTGSQVTFGFASTTLSNEE158 pKa = 3.52IFKK161 pKa = 10.93QPTSPGGGVVSVEE174 pKa = 4.16TGLPGAPTEE183 pKa = 4.44TVTGVSGNTASVSVAQQTVNLTGPAGATVRR213 pKa = 11.84VMVLEE218 pKa = 4.6GAAYY222 pKa = 7.37TQGVPSTSPQLGPYY236 pKa = 8.32QANSAVAVATYY247 pKa = 10.83DD248 pKa = 3.41VTLDD252 pKa = 3.88GQGHH256 pKa = 6.85GSTPVTLTHH265 pKa = 6.39TVTEE269 pKa = 4.45GGLNIITSAVVGADD283 pKa = 3.19GVVGPASDD291 pKa = 5.64PITLAYY297 pKa = 9.98NVSPPPPPPPPPPVAGVTIVQSGGSTAVTEE327 pKa = 4.71GGATDD332 pKa = 3.87TVTLALAKK340 pKa = 10.44QPTANVTVTVAGDD353 pKa = 3.66ADD355 pKa = 4.02VTGAPQSFTFTPTNWSTPQTLTVTAVDD382 pKa = 3.55DD383 pKa = 4.03TFIEE387 pKa = 4.72GTEE390 pKa = 4.03TAHH393 pKa = 7.57LGFTTASPDD402 pKa = 3.09TSYY405 pKa = 11.95NNLAVAPLNVTVTDD419 pKa = 4.06NDD421 pKa = 3.4QSSTPNFVQSQIAGLPDD438 pKa = 3.53ANYY441 pKa = 9.88TSLQFGPDD449 pKa = 2.79GRR451 pKa = 11.84LYY453 pKa = 10.53VSKK456 pKa = 11.2VDD458 pKa = 3.72GTILALKK465 pKa = 9.35LQQTTTGYY473 pKa = 9.99QVAAGGIEE481 pKa = 4.26TINAIKK487 pKa = 10.61SITNYY492 pKa = 10.62NDD494 pKa = 3.78DD495 pKa = 3.89GTLASAGYY503 pKa = 6.93QTSRR507 pKa = 11.84QVTGIFVTSGAAPAGGGAAPVVMYY531 pKa = 10.11VSSSDD536 pKa = 3.34PRR538 pKa = 11.84IGAGVNGGDD547 pKa = 4.0VNLDD551 pKa = 3.59TNSGVISRR559 pKa = 11.84LTQQTDD565 pKa = 3.88GSWQRR570 pKa = 11.84VDD572 pKa = 4.07LVRR575 pKa = 11.84GLPRR579 pKa = 11.84SEE581 pKa = 4.58EE582 pKa = 3.91NHH584 pKa = 6.08SVNGLQVTPDD594 pKa = 3.36GTTMYY599 pKa = 10.62LAIGGNTNMGAPSNNFAYY617 pKa = 9.1TPEE620 pKa = 4.12YY621 pKa = 10.28AYY623 pKa = 10.86SAAILKK629 pKa = 9.83IDD631 pKa = 4.62LVAIDD636 pKa = 4.72AMASKK641 pKa = 9.72TDD643 pKa = 3.53QFGNTYY649 pKa = 10.35KK650 pKa = 11.01YY651 pKa = 10.97NLPTLPASAGDD662 pKa = 4.78PYY664 pKa = 10.92PSDD667 pKa = 3.68PFGGSDD673 pKa = 3.65GFSQAKK679 pKa = 8.89IVAGSPVQIFSPGYY693 pKa = 8.38RR694 pKa = 11.84NPYY697 pKa = 10.43DD698 pKa = 3.93LVLTQSGQIYY708 pKa = 8.42TVDD711 pKa = 3.1NGANAGWGGVPVNEE725 pKa = 4.23GTPNVTNAPVEE736 pKa = 4.33GGVDD740 pKa = 3.57VPDD743 pKa = 3.45QLFLVTQGFYY753 pKa = 10.75GGHH756 pKa = 6.98PNPIRR761 pKa = 11.84ANPTGAGLYY770 pKa = 9.6PNGSTTPTTTYY781 pKa = 9.12PQGWPPVSTADD792 pKa = 3.91PVEE795 pKa = 4.2GDD797 pKa = 3.37YY798 pKa = 11.05RR799 pKa = 11.84GPGQSGSLYY808 pKa = 10.81SGFQASTNGITEE820 pKa = 4.17YY821 pKa = 10.1TASTFGNAMKK831 pKa = 10.77GNLLIVSWDD840 pKa = 3.48DD841 pKa = 3.08QLYY844 pKa = 9.73RR845 pKa = 11.84VQLGANGTSVTSVTKK860 pKa = 9.3LTNGSTVLGNGSPLDD875 pKa = 3.63VTSPGDD881 pKa = 3.66GQPFAGNIFVANYY894 pKa = 10.08GGGITAFIPTASPPPPPPPPNVADD918 pKa = 3.68SDD920 pKa = 3.95GDD922 pKa = 4.31GIVDD926 pKa = 3.52TRR928 pKa = 11.84DD929 pKa = 3.29PFAVDD934 pKa = 3.82ASNGTATTVSAGQTLTWEE952 pKa = 5.17FSQNIAPPSPPGLFNMGFTGLMSNGVDD979 pKa = 4.19GYY981 pKa = 9.39TALYY985 pKa = 10.31DD986 pKa = 3.68PANIKK991 pKa = 10.35PGGAASGFQVEE1002 pKa = 4.64KK1003 pKa = 9.98TGSGDD1008 pKa = 3.1AHH1010 pKa = 7.18LDD1012 pKa = 3.3NHH1014 pKa = 6.27QEE1016 pKa = 3.74NGFQFGVHH1024 pKa = 6.17LQPGVTGVVFQTTIDD1039 pKa = 3.62NPFAGTPSTSASASKK1054 pKa = 10.59SQGFFIGTGDD1064 pKa = 3.28QDD1066 pKa = 3.64NYY1068 pKa = 10.8LKK1070 pKa = 10.93VVATGAQGSGNIEE1083 pKa = 3.75VDD1085 pKa = 3.55TEE1087 pKa = 4.25NAGVWASTAYY1097 pKa = 9.51KK1098 pKa = 10.72VNIISNSVGALDD1110 pKa = 5.11TITLRR1115 pKa = 11.84LNVDD1119 pKa = 3.41VTTGLATPSWSYY1131 pKa = 9.6TVNGVASTGSGAAVQLSGATLSALNGAYY1159 pKa = 8.95TVDD1162 pKa = 3.41GHH1164 pKa = 7.11ASALAVGIISTSDD1177 pKa = 3.12GTNDD1181 pKa = 3.47PFTALWQNMSITASGSGGSPPPPPPPPPPPPPPPPPPPPPPSGAGVVLTATAGGSNLVGGSGDD1244 pKa = 3.54DD1245 pKa = 3.64TLVGGSGADD1254 pKa = 3.21TMTGAGGADD1263 pKa = 3.09HH1264 pKa = 7.12FVFPHH1269 pKa = 6.56LAWTSDD1275 pKa = 3.02QVTDD1279 pKa = 4.07FTDD1282 pKa = 3.91GTDD1285 pKa = 4.17LIDD1288 pKa = 3.66VTGLLSDD1295 pKa = 4.07AGYY1298 pKa = 8.93TGSNPIADD1306 pKa = 4.42GYY1308 pKa = 11.16VKK1310 pKa = 10.84LIANGSGGTWLYY1322 pKa = 10.78FDD1324 pKa = 5.11RR1325 pKa = 11.84DD1326 pKa = 3.68GTGTADD1332 pKa = 2.79QWGTYY1337 pKa = 7.15VANISGVTPSQLTSADD1353 pKa = 3.79FVASGSPPPPPSPPPPPPPPPPPPPPPPPPPPPPPPPPPPPSGGGVVLTASAGGSNLVGGSGDD1416 pKa = 3.54DD1417 pKa = 3.64TLVGGSGADD1426 pKa = 3.21TMTGAGGADD1435 pKa = 3.09HH1436 pKa = 7.12FVFPHH1441 pKa = 6.56LAWTSDD1447 pKa = 3.02QVTDD1451 pKa = 4.07FTDD1454 pKa = 3.91GTDD1457 pKa = 4.17LIDD1460 pKa = 3.66VTGLLSDD1467 pKa = 4.07AGYY1470 pKa = 8.93TGSNPIADD1478 pKa = 4.42GYY1480 pKa = 11.16VKK1482 pKa = 10.84LIANGSGGTWLYY1494 pKa = 10.78FDD1496 pKa = 5.11RR1497 pKa = 11.84DD1498 pKa = 3.68GTGTADD1504 pKa = 2.79QWGTYY1509 pKa = 7.15VANISGVTPTQLTSADD1525 pKa = 3.97FVASGSPPPPPSPPPPPPPPPPPPPPPPPPPPPPPPPSGGVVLTATAGGSNLVGGAGDD1583 pKa = 3.66DD1584 pKa = 3.78TLNAGSGPDD1593 pKa = 3.4TLTGGGGADD1602 pKa = 3.08HH1603 pKa = 6.43MVWGDD1608 pKa = 3.77VPWTAGHH1615 pKa = 5.6VTDD1618 pKa = 5.07FTLGADD1624 pKa = 3.93KK1625 pKa = 11.05LDD1627 pKa = 3.8AKK1629 pKa = 10.92AWLAKK1634 pKa = 10.14VGYY1637 pKa = 10.42AGTNPIADD1645 pKa = 3.75QYY1647 pKa = 11.46IKK1649 pKa = 10.62LISDD1653 pKa = 4.1GAGNTWVYY1661 pKa = 10.6FDD1663 pKa = 5.13RR1664 pKa = 11.84DD1665 pKa = 3.58GTGTADD1671 pKa = 2.63KK1672 pKa = 10.06WGTFVTTLDD1681 pKa = 3.42HH1682 pKa = 6.11VAPGSITLGDD1692 pKa = 4.32WIIKK1696 pKa = 9.51
MM1 pKa = 7.86PSGPSGSLLVTPGGPITDD19 pKa = 3.4STYY22 pKa = 8.35TARR25 pKa = 11.84SFRR28 pKa = 11.84LTNTSTDD35 pKa = 3.44GQQLNTVTIDD45 pKa = 2.91ISGSVLGGMIYY56 pKa = 10.59DD57 pKa = 3.86PTGQAGDD64 pKa = 3.51STARR68 pKa = 11.84PFTIDD73 pKa = 3.09SQTGTFSLVNATLSNGSDD91 pKa = 2.86ATGYY95 pKa = 6.68TTLTLQLSGFDD106 pKa = 3.89PGEE109 pKa = 4.06EE110 pKa = 4.15LDD112 pKa = 4.53FSIDD116 pKa = 3.17VDD118 pKa = 4.01PNSIRR123 pKa = 11.84GTTSPGPGEE132 pKa = 4.39SGSVAGVEE140 pKa = 4.23LTGSQVTFGFASTTLSNEE158 pKa = 3.52IFKK161 pKa = 10.93QPTSPGGGVVSVEE174 pKa = 4.16TGLPGAPTEE183 pKa = 4.44TVTGVSGNTASVSVAQQTVNLTGPAGATVRR213 pKa = 11.84VMVLEE218 pKa = 4.6GAAYY222 pKa = 7.37TQGVPSTSPQLGPYY236 pKa = 8.32QANSAVAVATYY247 pKa = 10.83DD248 pKa = 3.41VTLDD252 pKa = 3.88GQGHH256 pKa = 6.85GSTPVTLTHH265 pKa = 6.39TVTEE269 pKa = 4.45GGLNIITSAVVGADD283 pKa = 3.19GVVGPASDD291 pKa = 5.64PITLAYY297 pKa = 9.98NVSPPPPPPPPPPVAGVTIVQSGGSTAVTEE327 pKa = 4.71GGATDD332 pKa = 3.87TVTLALAKK340 pKa = 10.44QPTANVTVTVAGDD353 pKa = 3.66ADD355 pKa = 4.02VTGAPQSFTFTPTNWSTPQTLTVTAVDD382 pKa = 3.55DD383 pKa = 4.03TFIEE387 pKa = 4.72GTEE390 pKa = 4.03TAHH393 pKa = 7.57LGFTTASPDD402 pKa = 3.09TSYY405 pKa = 11.95NNLAVAPLNVTVTDD419 pKa = 4.06NDD421 pKa = 3.4QSSTPNFVQSQIAGLPDD438 pKa = 3.53ANYY441 pKa = 9.88TSLQFGPDD449 pKa = 2.79GRR451 pKa = 11.84LYY453 pKa = 10.53VSKK456 pKa = 11.2VDD458 pKa = 3.72GTILALKK465 pKa = 9.35LQQTTTGYY473 pKa = 9.99QVAAGGIEE481 pKa = 4.26TINAIKK487 pKa = 10.61SITNYY492 pKa = 10.62NDD494 pKa = 3.78DD495 pKa = 3.89GTLASAGYY503 pKa = 6.93QTSRR507 pKa = 11.84QVTGIFVTSGAAPAGGGAAPVVMYY531 pKa = 10.11VSSSDD536 pKa = 3.34PRR538 pKa = 11.84IGAGVNGGDD547 pKa = 4.0VNLDD551 pKa = 3.59TNSGVISRR559 pKa = 11.84LTQQTDD565 pKa = 3.88GSWQRR570 pKa = 11.84VDD572 pKa = 4.07LVRR575 pKa = 11.84GLPRR579 pKa = 11.84SEE581 pKa = 4.58EE582 pKa = 3.91NHH584 pKa = 6.08SVNGLQVTPDD594 pKa = 3.36GTTMYY599 pKa = 10.62LAIGGNTNMGAPSNNFAYY617 pKa = 9.1TPEE620 pKa = 4.12YY621 pKa = 10.28AYY623 pKa = 10.86SAAILKK629 pKa = 9.83IDD631 pKa = 4.62LVAIDD636 pKa = 4.72AMASKK641 pKa = 9.72TDD643 pKa = 3.53QFGNTYY649 pKa = 10.35KK650 pKa = 11.01YY651 pKa = 10.97NLPTLPASAGDD662 pKa = 4.78PYY664 pKa = 10.92PSDD667 pKa = 3.68PFGGSDD673 pKa = 3.65GFSQAKK679 pKa = 8.89IVAGSPVQIFSPGYY693 pKa = 8.38RR694 pKa = 11.84NPYY697 pKa = 10.43DD698 pKa = 3.93LVLTQSGQIYY708 pKa = 8.42TVDD711 pKa = 3.1NGANAGWGGVPVNEE725 pKa = 4.23GTPNVTNAPVEE736 pKa = 4.33GGVDD740 pKa = 3.57VPDD743 pKa = 3.45QLFLVTQGFYY753 pKa = 10.75GGHH756 pKa = 6.98PNPIRR761 pKa = 11.84ANPTGAGLYY770 pKa = 9.6PNGSTTPTTTYY781 pKa = 9.12PQGWPPVSTADD792 pKa = 3.91PVEE795 pKa = 4.2GDD797 pKa = 3.37YY798 pKa = 11.05RR799 pKa = 11.84GPGQSGSLYY808 pKa = 10.81SGFQASTNGITEE820 pKa = 4.17YY821 pKa = 10.1TASTFGNAMKK831 pKa = 10.77GNLLIVSWDD840 pKa = 3.48DD841 pKa = 3.08QLYY844 pKa = 9.73RR845 pKa = 11.84VQLGANGTSVTSVTKK860 pKa = 9.3LTNGSTVLGNGSPLDD875 pKa = 3.63VTSPGDD881 pKa = 3.66GQPFAGNIFVANYY894 pKa = 10.08GGGITAFIPTASPPPPPPPPNVADD918 pKa = 3.68SDD920 pKa = 3.95GDD922 pKa = 4.31GIVDD926 pKa = 3.52TRR928 pKa = 11.84DD929 pKa = 3.29PFAVDD934 pKa = 3.82ASNGTATTVSAGQTLTWEE952 pKa = 5.17FSQNIAPPSPPGLFNMGFTGLMSNGVDD979 pKa = 4.19GYY981 pKa = 9.39TALYY985 pKa = 10.31DD986 pKa = 3.68PANIKK991 pKa = 10.35PGGAASGFQVEE1002 pKa = 4.64KK1003 pKa = 9.98TGSGDD1008 pKa = 3.1AHH1010 pKa = 7.18LDD1012 pKa = 3.3NHH1014 pKa = 6.27QEE1016 pKa = 3.74NGFQFGVHH1024 pKa = 6.17LQPGVTGVVFQTTIDD1039 pKa = 3.62NPFAGTPSTSASASKK1054 pKa = 10.59SQGFFIGTGDD1064 pKa = 3.28QDD1066 pKa = 3.64NYY1068 pKa = 10.8LKK1070 pKa = 10.93VVATGAQGSGNIEE1083 pKa = 3.75VDD1085 pKa = 3.55TEE1087 pKa = 4.25NAGVWASTAYY1097 pKa = 9.51KK1098 pKa = 10.72VNIISNSVGALDD1110 pKa = 5.11TITLRR1115 pKa = 11.84LNVDD1119 pKa = 3.41VTTGLATPSWSYY1131 pKa = 9.6TVNGVASTGSGAAVQLSGATLSALNGAYY1159 pKa = 8.95TVDD1162 pKa = 3.41GHH1164 pKa = 7.11ASALAVGIISTSDD1177 pKa = 3.12GTNDD1181 pKa = 3.47PFTALWQNMSITASGSGGSPPPPPPPPPPPPPPPPPPPPPPSGAGVVLTATAGGSNLVGGSGDD1244 pKa = 3.54DD1245 pKa = 3.64TLVGGSGADD1254 pKa = 3.21TMTGAGGADD1263 pKa = 3.09HH1264 pKa = 7.12FVFPHH1269 pKa = 6.56LAWTSDD1275 pKa = 3.02QVTDD1279 pKa = 4.07FTDD1282 pKa = 3.91GTDD1285 pKa = 4.17LIDD1288 pKa = 3.66VTGLLSDD1295 pKa = 4.07AGYY1298 pKa = 8.93TGSNPIADD1306 pKa = 4.42GYY1308 pKa = 11.16VKK1310 pKa = 10.84LIANGSGGTWLYY1322 pKa = 10.78FDD1324 pKa = 5.11RR1325 pKa = 11.84DD1326 pKa = 3.68GTGTADD1332 pKa = 2.79QWGTYY1337 pKa = 7.15VANISGVTPSQLTSADD1353 pKa = 3.79FVASGSPPPPPSPPPPPPPPPPPPPPPPPPPPPPPPPPPPPSGGGVVLTASAGGSNLVGGSGDD1416 pKa = 3.54DD1417 pKa = 3.64TLVGGSGADD1426 pKa = 3.21TMTGAGGADD1435 pKa = 3.09HH1436 pKa = 7.12FVFPHH1441 pKa = 6.56LAWTSDD1447 pKa = 3.02QVTDD1451 pKa = 4.07FTDD1454 pKa = 3.91GTDD1457 pKa = 4.17LIDD1460 pKa = 3.66VTGLLSDD1467 pKa = 4.07AGYY1470 pKa = 8.93TGSNPIADD1478 pKa = 4.42GYY1480 pKa = 11.16VKK1482 pKa = 10.84LIANGSGGTWLYY1494 pKa = 10.78FDD1496 pKa = 5.11RR1497 pKa = 11.84DD1498 pKa = 3.68GTGTADD1504 pKa = 2.79QWGTYY1509 pKa = 7.15VANISGVTPTQLTSADD1525 pKa = 3.97FVASGSPPPPPSPPPPPPPPPPPPPPPPPPPPPPPPPSGGVVLTATAGGSNLVGGAGDD1583 pKa = 3.66DD1584 pKa = 3.78TLNAGSGPDD1593 pKa = 3.4TLTGGGGADD1602 pKa = 3.08HH1603 pKa = 6.43MVWGDD1608 pKa = 3.77VPWTAGHH1615 pKa = 5.6VTDD1618 pKa = 5.07FTLGADD1624 pKa = 3.93KK1625 pKa = 11.05LDD1627 pKa = 3.8AKK1629 pKa = 10.92AWLAKK1634 pKa = 10.14VGYY1637 pKa = 10.42AGTNPIADD1645 pKa = 3.75QYY1647 pKa = 11.46IKK1649 pKa = 10.62LISDD1653 pKa = 4.1GAGNTWVYY1661 pKa = 10.6FDD1663 pKa = 5.13RR1664 pKa = 11.84DD1665 pKa = 3.58GTGTADD1671 pKa = 2.63KK1672 pKa = 10.06WGTFVTTLDD1681 pKa = 3.42HH1682 pKa = 6.11VAPGSITLGDD1692 pKa = 4.32WIIKK1696 pKa = 9.51
Molecular weight: 170.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A328AGA1|A0A328AGA1_9CAUL Uncharacterized protein OS=Phenylobacterium soli OX=2170551 GN=DJ017_00610 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.11RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.53GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTKK25 pKa = 10.17NGQKK29 pKa = 9.5VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.11RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.53GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTKK25 pKa = 10.17NGQKK29 pKa = 9.5VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1208133 |
14 |
2196 |
309.4 |
33.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.292 ± 0.072 | 0.785 ± 0.012 |
5.572 ± 0.031 | 5.862 ± 0.044 |
3.495 ± 0.022 | 9.062 ± 0.042 |
1.928 ± 0.016 | 4.107 ± 0.027 |
3.176 ± 0.034 | 9.968 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.235 ± 0.021 | 2.225 ± 0.03 |
5.715 ± 0.033 | 3.181 ± 0.024 |
7.443 ± 0.043 | 4.795 ± 0.032 |
5.035 ± 0.047 | 7.491 ± 0.028 |
1.447 ± 0.016 | 2.187 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
