
Beggiatoa sp. PS
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; Thiotrichaceae; Beggiatoa; unclassified Beggiatoa
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
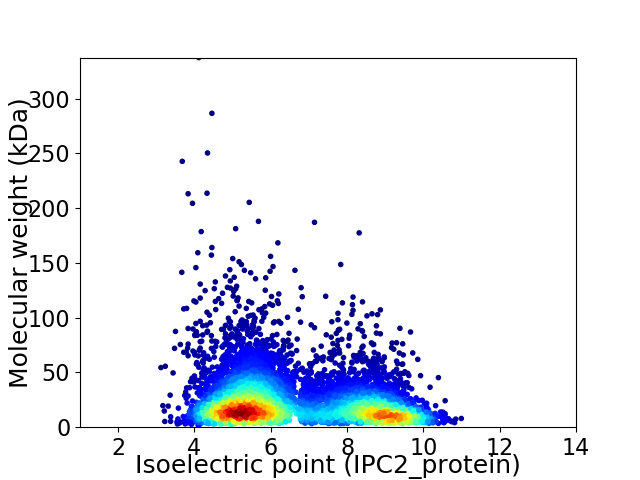
Virtual 2D-PAGE plot for 6644 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A7BSP7|A7BSP7_9GAMM Uncharacterized protein OS=Beggiatoa sp. PS OX=422289 GN=BGP_1754 PE=4 SV=1
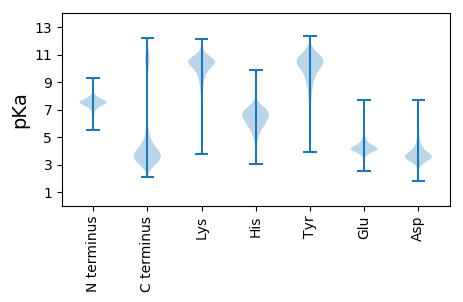
MM1 pKa = 7.94DD2 pKa = 4.56GEE4 pKa = 4.59EE5 pKa = 4.46VEE7 pKa = 5.13SSTTLPLEE15 pKa = 4.74GGLHH19 pKa = 5.93SIKK22 pKa = 10.4VEE24 pKa = 4.21YY25 pKa = 10.34VAGTSITVDD34 pKa = 2.66WSEE37 pKa = 3.98TSEE40 pKa = 3.87IAGNNLSFDD49 pKa = 3.61GMDD52 pKa = 4.8DD53 pKa = 3.95YY54 pKa = 12.21VDD56 pKa = 3.89LQTDD60 pKa = 3.94VVEE63 pKa = 4.41LAQADD68 pKa = 4.21FSLEE72 pKa = 3.58AWINTAYY79 pKa = 10.47SGSGQGILVSNDD91 pKa = 3.13ADD93 pKa = 4.02SSWEE97 pKa = 3.82LGEE100 pKa = 4.33KK101 pKa = 10.15AFHH104 pKa = 5.63VTSSGTLQFVGWGNGYY120 pKa = 9.92IKK122 pKa = 10.8GSTAVNDD129 pKa = 4.81GQWHH133 pKa = 5.53HH134 pKa = 6.42VAVVWDD140 pKa = 3.89YY141 pKa = 10.14TAGSGKK147 pKa = 10.02IYY149 pKa = 10.59LDD151 pKa = 3.94GADD154 pKa = 4.1DD155 pKa = 4.09TDD157 pKa = 3.68TGGTNYY163 pKa = 9.89VASNANNAGDD173 pKa = 4.04TLKK176 pKa = 10.45IAYY179 pKa = 10.3SNFNSSEE186 pKa = 3.72APNYY190 pKa = 9.98FNGGIDD196 pKa = 4.0EE197 pKa = 4.2VRR199 pKa = 11.84VWNTARR205 pKa = 11.84SQAEE209 pKa = 3.45IQAYY213 pKa = 7.74MKK215 pKa = 9.76STLKK219 pKa = 9.88GTEE222 pKa = 4.21SNLVAYY228 pKa = 8.59YY229 pKa = 10.43QLDD232 pKa = 3.87EE233 pKa = 4.64SSGTTATDD241 pKa = 3.15TTGNYY246 pKa = 10.39DD247 pKa = 3.23GTLNGDD253 pKa = 3.86PTWGVSDD260 pKa = 5.28APLTDD265 pKa = 4.26LPAPGSALEE274 pKa = 4.42LDD276 pKa = 3.73GSGDD280 pKa = 4.54YY281 pKa = 9.24ITLPDD286 pKa = 4.46DD287 pKa = 3.7KK288 pKa = 10.86TWFNGDD294 pKa = 3.43LTIEE298 pKa = 4.17SWVYY302 pKa = 8.04PHH304 pKa = 7.3SYY306 pKa = 10.98ANWSRR311 pKa = 11.84ILDD314 pKa = 3.68IGQGSGDD321 pKa = 3.93YY322 pKa = 10.8NVVLALTNEE331 pKa = 4.44TSGKK335 pKa = 9.02PSFRR339 pKa = 11.84VFDD342 pKa = 4.55DD343 pKa = 3.42NGNRR347 pKa = 11.84NPLEE351 pKa = 4.43SNTALTISTWNHH363 pKa = 4.82IAVTLSGTTATMYY376 pKa = 10.81INGAEE381 pKa = 4.24SGSITNSYY389 pKa = 9.62IPSDD393 pKa = 3.67VTRR396 pKa = 11.84DD397 pKa = 3.2KK398 pKa = 11.45AYY400 pKa = 9.7IGHH403 pKa = 6.19SNWSNDD409 pKa = 2.49ADD411 pKa = 3.79ANAVFDD417 pKa = 5.1EE418 pKa = 3.93MRR420 pKa = 11.84IWDD423 pKa = 3.83VARR426 pKa = 11.84SQAEE430 pKa = 3.45IQAYY434 pKa = 7.31MNRR437 pKa = 11.84TMDD440 pKa = 3.63GNEE443 pKa = 4.15SGLIAYY449 pKa = 9.03YY450 pKa = 10.94NFDD453 pKa = 3.63EE454 pKa = 4.9KK455 pKa = 10.91AASSIAVLSDD465 pKa = 2.93IAGNNLGTLNGNPTWVDD482 pKa = 2.39SDD484 pKa = 4.27APVSKK489 pKa = 10.23MIAPGNAMSFDD500 pKa = 3.69GVDD503 pKa = 3.79DD504 pKa = 3.87YY505 pKa = 12.18VAFDD509 pKa = 3.88DD510 pKa = 5.61TIPLDD515 pKa = 3.53DD516 pKa = 4.35TYY518 pKa = 11.08TIEE521 pKa = 4.93GWFQTSNTGDD531 pKa = 3.17ILSFTDD537 pKa = 3.52SGNHH541 pKa = 4.88YY542 pKa = 11.05ALIEE546 pKa = 3.94LAGNGIMRR554 pKa = 11.84FLHH557 pKa = 6.08RR558 pKa = 11.84VPAGSANGTNIYY570 pKa = 10.53SSAGYY575 pKa = 10.91ADD577 pKa = 4.56GSWHH581 pKa = 6.91HH582 pKa = 6.23FACVKK587 pKa = 10.76DD588 pKa = 3.63EE589 pKa = 4.14TGMYY593 pKa = 9.69IYY595 pKa = 10.09MDD597 pKa = 4.33GEE599 pKa = 4.5QVASGSDD606 pKa = 3.39SNAIGDD612 pKa = 4.03SVEE615 pKa = 4.13VVIGRR620 pKa = 11.84LSPTNSSRR628 pKa = 11.84YY629 pKa = 9.25FEE631 pKa = 4.74GSLDD635 pKa = 4.79EE636 pKa = 3.91IRR638 pKa = 11.84IWNTARR644 pKa = 11.84TQTEE648 pKa = 3.9IQADD652 pKa = 4.07MYY654 pKa = 10.64RR655 pKa = 11.84ALGSFEE661 pKa = 4.3TGLVAYY667 pKa = 9.45YY668 pKa = 10.72KK669 pKa = 10.33FDD671 pKa = 3.52QDD673 pKa = 3.33SGTEE677 pKa = 4.17VVDD680 pKa = 3.15ITSFDD685 pKa = 3.53NDD687 pKa = 3.14GTLNGGPSWVTSEE700 pKa = 4.37VPMDD704 pKa = 3.59
MM1 pKa = 7.94DD2 pKa = 4.56GEE4 pKa = 4.59EE5 pKa = 4.46VEE7 pKa = 5.13SSTTLPLEE15 pKa = 4.74GGLHH19 pKa = 5.93SIKK22 pKa = 10.4VEE24 pKa = 4.21YY25 pKa = 10.34VAGTSITVDD34 pKa = 2.66WSEE37 pKa = 3.98TSEE40 pKa = 3.87IAGNNLSFDD49 pKa = 3.61GMDD52 pKa = 4.8DD53 pKa = 3.95YY54 pKa = 12.21VDD56 pKa = 3.89LQTDD60 pKa = 3.94VVEE63 pKa = 4.41LAQADD68 pKa = 4.21FSLEE72 pKa = 3.58AWINTAYY79 pKa = 10.47SGSGQGILVSNDD91 pKa = 3.13ADD93 pKa = 4.02SSWEE97 pKa = 3.82LGEE100 pKa = 4.33KK101 pKa = 10.15AFHH104 pKa = 5.63VTSSGTLQFVGWGNGYY120 pKa = 9.92IKK122 pKa = 10.8GSTAVNDD129 pKa = 4.81GQWHH133 pKa = 5.53HH134 pKa = 6.42VAVVWDD140 pKa = 3.89YY141 pKa = 10.14TAGSGKK147 pKa = 10.02IYY149 pKa = 10.59LDD151 pKa = 3.94GADD154 pKa = 4.1DD155 pKa = 4.09TDD157 pKa = 3.68TGGTNYY163 pKa = 9.89VASNANNAGDD173 pKa = 4.04TLKK176 pKa = 10.45IAYY179 pKa = 10.3SNFNSSEE186 pKa = 3.72APNYY190 pKa = 9.98FNGGIDD196 pKa = 4.0EE197 pKa = 4.2VRR199 pKa = 11.84VWNTARR205 pKa = 11.84SQAEE209 pKa = 3.45IQAYY213 pKa = 7.74MKK215 pKa = 9.76STLKK219 pKa = 9.88GTEE222 pKa = 4.21SNLVAYY228 pKa = 8.59YY229 pKa = 10.43QLDD232 pKa = 3.87EE233 pKa = 4.64SSGTTATDD241 pKa = 3.15TTGNYY246 pKa = 10.39DD247 pKa = 3.23GTLNGDD253 pKa = 3.86PTWGVSDD260 pKa = 5.28APLTDD265 pKa = 4.26LPAPGSALEE274 pKa = 4.42LDD276 pKa = 3.73GSGDD280 pKa = 4.54YY281 pKa = 9.24ITLPDD286 pKa = 4.46DD287 pKa = 3.7KK288 pKa = 10.86TWFNGDD294 pKa = 3.43LTIEE298 pKa = 4.17SWVYY302 pKa = 8.04PHH304 pKa = 7.3SYY306 pKa = 10.98ANWSRR311 pKa = 11.84ILDD314 pKa = 3.68IGQGSGDD321 pKa = 3.93YY322 pKa = 10.8NVVLALTNEE331 pKa = 4.44TSGKK335 pKa = 9.02PSFRR339 pKa = 11.84VFDD342 pKa = 4.55DD343 pKa = 3.42NGNRR347 pKa = 11.84NPLEE351 pKa = 4.43SNTALTISTWNHH363 pKa = 4.82IAVTLSGTTATMYY376 pKa = 10.81INGAEE381 pKa = 4.24SGSITNSYY389 pKa = 9.62IPSDD393 pKa = 3.67VTRR396 pKa = 11.84DD397 pKa = 3.2KK398 pKa = 11.45AYY400 pKa = 9.7IGHH403 pKa = 6.19SNWSNDD409 pKa = 2.49ADD411 pKa = 3.79ANAVFDD417 pKa = 5.1EE418 pKa = 3.93MRR420 pKa = 11.84IWDD423 pKa = 3.83VARR426 pKa = 11.84SQAEE430 pKa = 3.45IQAYY434 pKa = 7.31MNRR437 pKa = 11.84TMDD440 pKa = 3.63GNEE443 pKa = 4.15SGLIAYY449 pKa = 9.03YY450 pKa = 10.94NFDD453 pKa = 3.63EE454 pKa = 4.9KK455 pKa = 10.91AASSIAVLSDD465 pKa = 2.93IAGNNLGTLNGNPTWVDD482 pKa = 2.39SDD484 pKa = 4.27APVSKK489 pKa = 10.23MIAPGNAMSFDD500 pKa = 3.69GVDD503 pKa = 3.79DD504 pKa = 3.87YY505 pKa = 12.18VAFDD509 pKa = 3.88DD510 pKa = 5.61TIPLDD515 pKa = 3.53DD516 pKa = 4.35TYY518 pKa = 11.08TIEE521 pKa = 4.93GWFQTSNTGDD531 pKa = 3.17ILSFTDD537 pKa = 3.52SGNHH541 pKa = 4.88YY542 pKa = 11.05ALIEE546 pKa = 3.94LAGNGIMRR554 pKa = 11.84FLHH557 pKa = 6.08RR558 pKa = 11.84VPAGSANGTNIYY570 pKa = 10.53SSAGYY575 pKa = 10.91ADD577 pKa = 4.56GSWHH581 pKa = 6.91HH582 pKa = 6.23FACVKK587 pKa = 10.76DD588 pKa = 3.63EE589 pKa = 4.14TGMYY593 pKa = 9.69IYY595 pKa = 10.09MDD597 pKa = 4.33GEE599 pKa = 4.5QVASGSDD606 pKa = 3.39SNAIGDD612 pKa = 4.03SVEE615 pKa = 4.13VVIGRR620 pKa = 11.84LSPTNSSRR628 pKa = 11.84YY629 pKa = 9.25FEE631 pKa = 4.74GSLDD635 pKa = 4.79EE636 pKa = 3.91IRR638 pKa = 11.84IWNTARR644 pKa = 11.84TQTEE648 pKa = 3.9IQADD652 pKa = 4.07MYY654 pKa = 10.64RR655 pKa = 11.84ALGSFEE661 pKa = 4.3TGLVAYY667 pKa = 9.45YY668 pKa = 10.72KK669 pKa = 10.33FDD671 pKa = 3.52QDD673 pKa = 3.33SGTEE677 pKa = 4.17VVDD680 pKa = 3.15ITSFDD685 pKa = 3.53NDD687 pKa = 3.14GTLNGGPSWVTSEE700 pKa = 4.37VPMDD704 pKa = 3.59
Molecular weight: 75.86 kDa
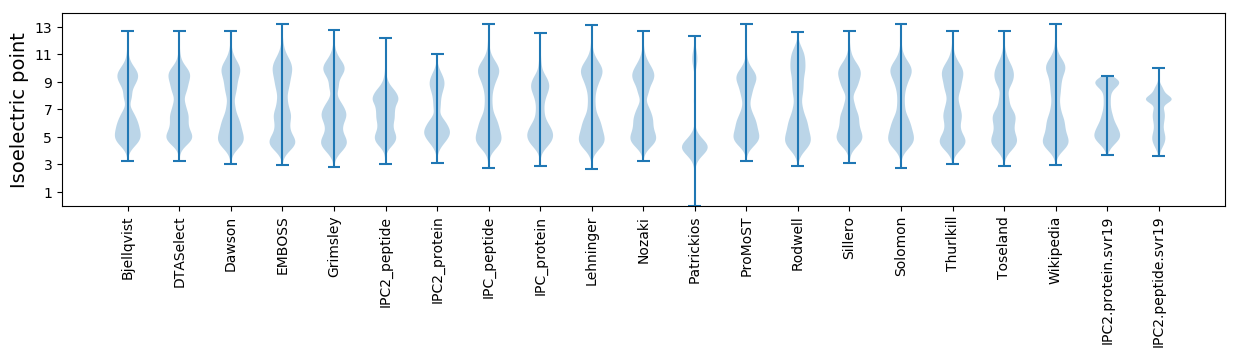
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7BUP4|A7BUP4_9GAMM Uncharacterized protein OS=Beggiatoa sp. PS OX=422289 GN=BGP_5125 PE=4 SV=1
MM1 pKa = 7.51TINRR5 pKa = 11.84LMAAKK10 pKa = 9.97PPMILKK16 pKa = 10.05RR17 pKa = 11.84QMALKK22 pKa = 10.05PLMILKK28 pKa = 9.64QRR30 pKa = 11.84MVLKK34 pKa = 10.49PPMILNRR41 pKa = 11.84RR42 pKa = 11.84MTINQLMALKK52 pKa = 9.29PQMMHH57 pKa = 6.63KK58 pKa = 10.13LKK60 pKa = 10.61HH61 pKa = 6.13RR62 pKa = 11.84SLLILL67 pKa = 3.99
MM1 pKa = 7.51TINRR5 pKa = 11.84LMAAKK10 pKa = 9.97PPMILKK16 pKa = 10.05RR17 pKa = 11.84QMALKK22 pKa = 10.05PLMILKK28 pKa = 9.64QRR30 pKa = 11.84MVLKK34 pKa = 10.49PPMILNRR41 pKa = 11.84RR42 pKa = 11.84MTINQLMALKK52 pKa = 9.29PQMMHH57 pKa = 6.63KK58 pKa = 10.13LKK60 pKa = 10.61HH61 pKa = 6.13RR62 pKa = 11.84SLLILL67 pKa = 3.99
Molecular weight: 7.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1440897 |
17 |
3115 |
216.9 |
24.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.802 ± 0.033 | 1.049 ± 0.013 |
5.095 ± 0.027 | 6.617 ± 0.037 |
4.476 ± 0.029 | 5.911 ± 0.041 |
2.33 ± 0.016 | 7.528 ± 0.032 |
6.24 ± 0.041 | 10.618 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.349 ± 0.017 | 4.931 ± 0.026 |
4.203 ± 0.025 | 5.056 ± 0.034 |
4.381 ± 0.028 | 6.031 ± 0.028 |
5.828 ± 0.035 | 5.868 ± 0.025 |
1.354 ± 0.015 | 3.335 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
