
Vibrio phage VvAW1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
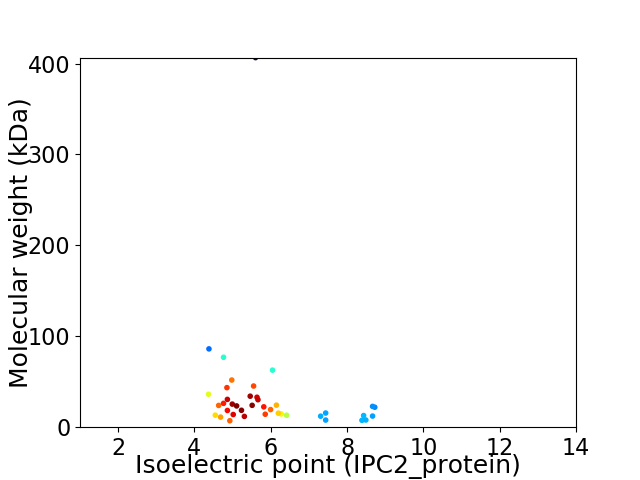
Virtual 2D-PAGE plot for 40 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3PV04|I3PV04_9CAUD SASA domain-containing protein OS=Vibrio phage VvAW1 OX=1168281 GN=VvAW1_00040c PE=4 SV=1
MM1 pKa = 7.69ANNLSMPRR9 pKa = 11.84FKK11 pKa = 11.17AFGQDD16 pKa = 3.38GKK18 pKa = 9.65PLAFGRR24 pKa = 11.84VYY26 pKa = 10.69LYY28 pKa = 10.55YY29 pKa = 10.2PRR31 pKa = 11.84TSEE34 pKa = 4.25KK35 pKa = 10.61KK36 pKa = 9.83PGYY39 pKa = 8.61TDD41 pKa = 2.93STANVEE47 pKa = 4.16LQNPVILDD55 pKa = 3.78ANGEE59 pKa = 4.0AAIFLDD65 pKa = 3.96GAYY68 pKa = 9.8KK69 pKa = 10.41IILRR73 pKa = 11.84DD74 pKa = 3.77KK75 pKa = 11.2NDD77 pKa = 3.31VLQWTVDD84 pKa = 3.43NYY86 pKa = 9.48NTDD89 pKa = 3.57YY90 pKa = 10.95FSSTNVSITPQANKK104 pKa = 10.02VPLAGSTGVITPGWIDD120 pKa = 3.53GLIAVYY126 pKa = 11.01NDD128 pKa = 3.03VAVSKK133 pKa = 11.31GDD135 pKa = 3.64IVLLKK140 pKa = 10.73QKK142 pKa = 10.43DD143 pKa = 3.74AEE145 pKa = 4.39HH146 pKa = 6.94DD147 pKa = 3.81TNISVLQTVTGDD159 pKa = 3.35NADD162 pKa = 4.7GITQLSIDD170 pKa = 3.77VQNLKK175 pKa = 10.4GASASLVKK183 pKa = 10.58GYY185 pKa = 8.32ATLAALNADD194 pKa = 4.79LVPEE198 pKa = 5.29DD199 pKa = 3.74GTLADD204 pKa = 3.91VTNDD208 pKa = 3.37PDD210 pKa = 3.9TNNNGRR216 pKa = 11.84YY217 pKa = 8.79RR218 pKa = 11.84KK219 pKa = 10.03SGATGTGSWVKK230 pKa = 9.82TNYY233 pKa = 10.19SEE235 pKa = 5.37WSTLWLEE242 pKa = 4.19MNRR245 pKa = 11.84LNDD248 pKa = 3.58FVYY251 pKa = 11.03DD252 pKa = 3.83LAIYY256 pKa = 10.0NYY258 pKa = 10.68NKK260 pKa = 8.75TQEE263 pKa = 4.2EE264 pKa = 4.69LEE266 pKa = 4.16IVPIVMDD273 pKa = 4.46EE274 pKa = 4.11ANKK277 pKa = 9.26MALGFYY283 pKa = 10.43SDD285 pKa = 4.29TGEE288 pKa = 3.73LWAAGIEE295 pKa = 4.2RR296 pKa = 11.84IEE298 pKa = 4.11ATGKK302 pKa = 9.67AVQPVVVNEE311 pKa = 4.32NGDD314 pKa = 3.38IAIGIEE320 pKa = 4.29SEE322 pKa = 4.23TGQTMMDD329 pKa = 3.35PHH331 pKa = 7.81YY332 pKa = 9.38STIMYY337 pKa = 9.08GVKK340 pKa = 10.11KK341 pKa = 10.77VGIFPDD347 pKa = 3.59EE348 pKa = 4.44FNVKK352 pKa = 10.1DD353 pKa = 4.05GYY355 pKa = 11.41VYY357 pKa = 10.83AIVDD361 pKa = 4.0EE362 pKa = 4.54DD363 pKa = 3.64KK364 pKa = 11.16KK365 pKa = 10.81IAFGIKK371 pKa = 9.84DD372 pKa = 3.91DD373 pKa = 4.06GSLVGSGLTFQTAPVEE389 pKa = 4.37KK390 pKa = 10.13GDD392 pKa = 3.72KK393 pKa = 10.25QSIDD397 pKa = 3.31MEE399 pKa = 4.75HH400 pKa = 6.03NHH402 pKa = 7.0FIFSGQSLSVGATGKK417 pKa = 9.7PVISTSQPYY426 pKa = 11.03DD427 pKa = 3.15NLTFDD432 pKa = 4.19GGPRR436 pKa = 11.84SDD438 pKa = 4.64LDD440 pKa = 3.57EE441 pKa = 4.31LTGFKK446 pKa = 10.5PLVEE450 pKa = 5.13DD451 pKa = 4.43EE452 pKa = 4.83NSPAPDD458 pKa = 3.49GGTNRR463 pKa = 11.84GEE465 pKa = 4.47TCCSGAANFTTHH477 pKa = 7.36LIEE480 pKa = 5.75SEE482 pKa = 4.16DD483 pKa = 4.02GIAYY487 pKa = 8.06DD488 pKa = 3.42QQGYY492 pKa = 10.37AIVSSTVGHH501 pKa = 6.15GGYY504 pKa = 10.36RR505 pKa = 11.84IDD507 pKa = 3.69QLYY510 pKa = 10.22KK511 pKa = 9.45GSAWYY516 pKa = 9.19DD517 pKa = 3.7YY518 pKa = 9.79YY519 pKa = 10.59TAHH522 pKa = 5.8VQAGFDD528 pKa = 3.59LSQAQGKK535 pKa = 7.12TYY537 pKa = 10.71AVQALGWIQGEE548 pKa = 3.8NDD550 pKa = 3.08QFDD553 pKa = 3.64NSKK556 pKa = 10.95ARR558 pKa = 11.84LAYY561 pKa = 9.89KK562 pKa = 10.35NDD564 pKa = 4.8LIQYY568 pKa = 8.51QADD571 pKa = 3.55IEE573 pKa = 4.66ADD575 pKa = 3.31AQAITGQTHH584 pKa = 6.31RR585 pKa = 11.84VPLLTYY591 pKa = 10.45QLACYY596 pKa = 9.26VRR598 pKa = 11.84QGYY601 pKa = 10.52RR602 pKa = 11.84NVTLAQLDD610 pKa = 4.06CHH612 pKa = 7.14KK613 pKa = 10.07EE614 pKa = 3.93SQDD617 pKa = 3.19IYY619 pKa = 11.01LVTPMYY625 pKa = 9.88HH626 pKa = 5.61VPYY629 pKa = 9.61ATDD632 pKa = 3.77AVHH635 pKa = 6.33LTNLGYY641 pKa = 10.84LWLGHH646 pKa = 5.74YY647 pKa = 9.53FGKK650 pKa = 10.58VYY652 pKa = 10.66KK653 pKa = 10.38KK654 pKa = 10.74VVFDD658 pKa = 4.55GIEE661 pKa = 3.91WEE663 pKa = 4.14PVRR666 pKa = 11.84PKK668 pKa = 10.54TITQQGAVVLIDD680 pKa = 4.27FFVPDD685 pKa = 5.09PPLQLDD691 pKa = 3.94TSTLGLATDD700 pKa = 4.14YY701 pKa = 11.54GFAAYY706 pKa = 9.78DD707 pKa = 3.49DD708 pKa = 4.6SGDD711 pKa = 3.46IDD713 pKa = 4.02VLSVEE718 pKa = 5.09LVGQTRR724 pKa = 11.84VKK726 pKa = 9.79LTLASAPGAGGGIRR740 pKa = 11.84YY741 pKa = 9.42GLDD744 pKa = 3.4YY745 pKa = 10.89IGTGLTHH752 pKa = 6.84NNGGSGNLRR761 pKa = 11.84DD762 pKa = 3.83SAPEE766 pKa = 3.78YY767 pKa = 9.29FTYY770 pKa = 10.81AGTDD774 pKa = 3.38YY775 pKa = 10.93PLYY778 pKa = 8.87NWCVMFDD785 pKa = 3.61EE786 pKa = 6.08LII788 pKa = 4.26
MM1 pKa = 7.69ANNLSMPRR9 pKa = 11.84FKK11 pKa = 11.17AFGQDD16 pKa = 3.38GKK18 pKa = 9.65PLAFGRR24 pKa = 11.84VYY26 pKa = 10.69LYY28 pKa = 10.55YY29 pKa = 10.2PRR31 pKa = 11.84TSEE34 pKa = 4.25KK35 pKa = 10.61KK36 pKa = 9.83PGYY39 pKa = 8.61TDD41 pKa = 2.93STANVEE47 pKa = 4.16LQNPVILDD55 pKa = 3.78ANGEE59 pKa = 4.0AAIFLDD65 pKa = 3.96GAYY68 pKa = 9.8KK69 pKa = 10.41IILRR73 pKa = 11.84DD74 pKa = 3.77KK75 pKa = 11.2NDD77 pKa = 3.31VLQWTVDD84 pKa = 3.43NYY86 pKa = 9.48NTDD89 pKa = 3.57YY90 pKa = 10.95FSSTNVSITPQANKK104 pKa = 10.02VPLAGSTGVITPGWIDD120 pKa = 3.53GLIAVYY126 pKa = 11.01NDD128 pKa = 3.03VAVSKK133 pKa = 11.31GDD135 pKa = 3.64IVLLKK140 pKa = 10.73QKK142 pKa = 10.43DD143 pKa = 3.74AEE145 pKa = 4.39HH146 pKa = 6.94DD147 pKa = 3.81TNISVLQTVTGDD159 pKa = 3.35NADD162 pKa = 4.7GITQLSIDD170 pKa = 3.77VQNLKK175 pKa = 10.4GASASLVKK183 pKa = 10.58GYY185 pKa = 8.32ATLAALNADD194 pKa = 4.79LVPEE198 pKa = 5.29DD199 pKa = 3.74GTLADD204 pKa = 3.91VTNDD208 pKa = 3.37PDD210 pKa = 3.9TNNNGRR216 pKa = 11.84YY217 pKa = 8.79RR218 pKa = 11.84KK219 pKa = 10.03SGATGTGSWVKK230 pKa = 9.82TNYY233 pKa = 10.19SEE235 pKa = 5.37WSTLWLEE242 pKa = 4.19MNRR245 pKa = 11.84LNDD248 pKa = 3.58FVYY251 pKa = 11.03DD252 pKa = 3.83LAIYY256 pKa = 10.0NYY258 pKa = 10.68NKK260 pKa = 8.75TQEE263 pKa = 4.2EE264 pKa = 4.69LEE266 pKa = 4.16IVPIVMDD273 pKa = 4.46EE274 pKa = 4.11ANKK277 pKa = 9.26MALGFYY283 pKa = 10.43SDD285 pKa = 4.29TGEE288 pKa = 3.73LWAAGIEE295 pKa = 4.2RR296 pKa = 11.84IEE298 pKa = 4.11ATGKK302 pKa = 9.67AVQPVVVNEE311 pKa = 4.32NGDD314 pKa = 3.38IAIGIEE320 pKa = 4.29SEE322 pKa = 4.23TGQTMMDD329 pKa = 3.35PHH331 pKa = 7.81YY332 pKa = 9.38STIMYY337 pKa = 9.08GVKK340 pKa = 10.11KK341 pKa = 10.77VGIFPDD347 pKa = 3.59EE348 pKa = 4.44FNVKK352 pKa = 10.1DD353 pKa = 4.05GYY355 pKa = 11.41VYY357 pKa = 10.83AIVDD361 pKa = 4.0EE362 pKa = 4.54DD363 pKa = 3.64KK364 pKa = 11.16KK365 pKa = 10.81IAFGIKK371 pKa = 9.84DD372 pKa = 3.91DD373 pKa = 4.06GSLVGSGLTFQTAPVEE389 pKa = 4.37KK390 pKa = 10.13GDD392 pKa = 3.72KK393 pKa = 10.25QSIDD397 pKa = 3.31MEE399 pKa = 4.75HH400 pKa = 6.03NHH402 pKa = 7.0FIFSGQSLSVGATGKK417 pKa = 9.7PVISTSQPYY426 pKa = 11.03DD427 pKa = 3.15NLTFDD432 pKa = 4.19GGPRR436 pKa = 11.84SDD438 pKa = 4.64LDD440 pKa = 3.57EE441 pKa = 4.31LTGFKK446 pKa = 10.5PLVEE450 pKa = 5.13DD451 pKa = 4.43EE452 pKa = 4.83NSPAPDD458 pKa = 3.49GGTNRR463 pKa = 11.84GEE465 pKa = 4.47TCCSGAANFTTHH477 pKa = 7.36LIEE480 pKa = 5.75SEE482 pKa = 4.16DD483 pKa = 4.02GIAYY487 pKa = 8.06DD488 pKa = 3.42QQGYY492 pKa = 10.37AIVSSTVGHH501 pKa = 6.15GGYY504 pKa = 10.36RR505 pKa = 11.84IDD507 pKa = 3.69QLYY510 pKa = 10.22KK511 pKa = 9.45GSAWYY516 pKa = 9.19DD517 pKa = 3.7YY518 pKa = 9.79YY519 pKa = 10.59TAHH522 pKa = 5.8VQAGFDD528 pKa = 3.59LSQAQGKK535 pKa = 7.12TYY537 pKa = 10.71AVQALGWIQGEE548 pKa = 3.8NDD550 pKa = 3.08QFDD553 pKa = 3.64NSKK556 pKa = 10.95ARR558 pKa = 11.84LAYY561 pKa = 9.89KK562 pKa = 10.35NDD564 pKa = 4.8LIQYY568 pKa = 8.51QADD571 pKa = 3.55IEE573 pKa = 4.66ADD575 pKa = 3.31AQAITGQTHH584 pKa = 6.31RR585 pKa = 11.84VPLLTYY591 pKa = 10.45QLACYY596 pKa = 9.26VRR598 pKa = 11.84QGYY601 pKa = 10.52RR602 pKa = 11.84NVTLAQLDD610 pKa = 4.06CHH612 pKa = 7.14KK613 pKa = 10.07EE614 pKa = 3.93SQDD617 pKa = 3.19IYY619 pKa = 11.01LVTPMYY625 pKa = 9.88HH626 pKa = 5.61VPYY629 pKa = 9.61ATDD632 pKa = 3.77AVHH635 pKa = 6.33LTNLGYY641 pKa = 10.84LWLGHH646 pKa = 5.74YY647 pKa = 9.53FGKK650 pKa = 10.58VYY652 pKa = 10.66KK653 pKa = 10.38KK654 pKa = 10.74VVFDD658 pKa = 4.55GIEE661 pKa = 3.91WEE663 pKa = 4.14PVRR666 pKa = 11.84PKK668 pKa = 10.54TITQQGAVVLIDD680 pKa = 4.27FFVPDD685 pKa = 5.09PPLQLDD691 pKa = 3.94TSTLGLATDD700 pKa = 4.14YY701 pKa = 11.54GFAAYY706 pKa = 9.78DD707 pKa = 3.49DD708 pKa = 4.6SGDD711 pKa = 3.46IDD713 pKa = 4.02VLSVEE718 pKa = 5.09LVGQTRR724 pKa = 11.84VKK726 pKa = 9.79LTLASAPGAGGGIRR740 pKa = 11.84YY741 pKa = 9.42GLDD744 pKa = 3.4YY745 pKa = 10.89IGTGLTHH752 pKa = 6.84NNGGSGNLRR761 pKa = 11.84DD762 pKa = 3.83SAPEE766 pKa = 3.78YY767 pKa = 9.29FTYY770 pKa = 10.81AGTDD774 pKa = 3.38YY775 pKa = 10.93PLYY778 pKa = 8.87NWCVMFDD785 pKa = 3.61EE786 pKa = 6.08LII788 pKa = 4.26
Molecular weight: 86.13 kDa
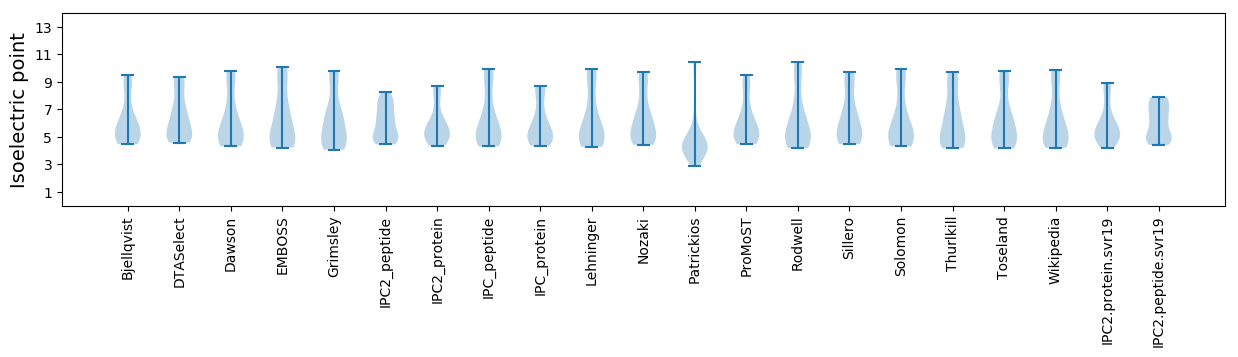
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3PUZ0|I3PUZ0_9CAUD RNA-binding regulator protein OS=Vibrio phage VvAW1 OX=1168281 GN=VvAW1_00026c PE=3 SV=1
MM1 pKa = 7.61AKK3 pKa = 10.11AKK5 pKa = 9.93PVDD8 pKa = 3.85VFGVRR13 pKa = 11.84YY14 pKa = 9.06KK15 pKa = 10.62SKK17 pKa = 10.12RR18 pKa = 11.84QACIAHH24 pKa = 5.93GQNPGKK30 pKa = 9.54IDD32 pKa = 3.23HH33 pKa = 7.19RR34 pKa = 11.84IAKK37 pKa = 9.9GMTLEE42 pKa = 4.85DD43 pKa = 3.32ALKK46 pKa = 10.96AKK48 pKa = 10.41DD49 pKa = 3.77RR50 pKa = 11.84YY51 pKa = 8.5TGAKK55 pKa = 8.61SHH57 pKa = 7.18PSYY60 pKa = 9.49THH62 pKa = 5.88WNGIRR67 pKa = 11.84SRR69 pKa = 11.84CLSDD73 pKa = 3.77SEE75 pKa = 4.21CSKK78 pKa = 10.37RR79 pKa = 11.84YY80 pKa = 9.83KK81 pKa = 10.7GVVDD85 pKa = 3.62ICDD88 pKa = 3.08RR89 pKa = 11.84WASDD93 pKa = 3.05FWSFVEE99 pKa = 5.98DD100 pKa = 3.11MGEE103 pKa = 3.98PPFVGATIEE112 pKa = 4.31RR113 pKa = 11.84LDD115 pKa = 3.52NSKK118 pKa = 11.07GYY120 pKa = 10.59DD121 pKa = 3.35KK122 pKa = 10.92EE123 pKa = 4.11NCIWASMKK131 pKa = 10.59DD132 pKa = 3.22QARR135 pKa = 11.84NRR137 pKa = 11.84RR138 pKa = 11.84TNVVIEE144 pKa = 4.4CFGEE148 pKa = 4.59TMCLADD154 pKa = 3.57WAVKK158 pKa = 9.63TGISSSLIAYY168 pKa = 7.94RR169 pKa = 11.84ISKK172 pKa = 9.33GWDD175 pKa = 2.89VEE177 pKa = 4.19RR178 pKa = 11.84ALTLKK183 pKa = 10.34PKK185 pKa = 10.31RR186 pKa = 11.84GRR188 pKa = 11.84NHH190 pKa = 5.57VNKK193 pKa = 10.5SS194 pKa = 3.03
MM1 pKa = 7.61AKK3 pKa = 10.11AKK5 pKa = 9.93PVDD8 pKa = 3.85VFGVRR13 pKa = 11.84YY14 pKa = 9.06KK15 pKa = 10.62SKK17 pKa = 10.12RR18 pKa = 11.84QACIAHH24 pKa = 5.93GQNPGKK30 pKa = 9.54IDD32 pKa = 3.23HH33 pKa = 7.19RR34 pKa = 11.84IAKK37 pKa = 9.9GMTLEE42 pKa = 4.85DD43 pKa = 3.32ALKK46 pKa = 10.96AKK48 pKa = 10.41DD49 pKa = 3.77RR50 pKa = 11.84YY51 pKa = 8.5TGAKK55 pKa = 8.61SHH57 pKa = 7.18PSYY60 pKa = 9.49THH62 pKa = 5.88WNGIRR67 pKa = 11.84SRR69 pKa = 11.84CLSDD73 pKa = 3.77SEE75 pKa = 4.21CSKK78 pKa = 10.37RR79 pKa = 11.84YY80 pKa = 9.83KK81 pKa = 10.7GVVDD85 pKa = 3.62ICDD88 pKa = 3.08RR89 pKa = 11.84WASDD93 pKa = 3.05FWSFVEE99 pKa = 5.98DD100 pKa = 3.11MGEE103 pKa = 3.98PPFVGATIEE112 pKa = 4.31RR113 pKa = 11.84LDD115 pKa = 3.52NSKK118 pKa = 11.07GYY120 pKa = 10.59DD121 pKa = 3.35KK122 pKa = 10.92EE123 pKa = 4.11NCIWASMKK131 pKa = 10.59DD132 pKa = 3.22QARR135 pKa = 11.84NRR137 pKa = 11.84RR138 pKa = 11.84TNVVIEE144 pKa = 4.4CFGEE148 pKa = 4.59TMCLADD154 pKa = 3.57WAVKK158 pKa = 9.63TGISSSLIAYY168 pKa = 7.94RR169 pKa = 11.84ISKK172 pKa = 9.33GWDD175 pKa = 2.89VEE177 pKa = 4.19RR178 pKa = 11.84ALTLKK183 pKa = 10.34PKK185 pKa = 10.31RR186 pKa = 11.84GRR188 pKa = 11.84NHH190 pKa = 5.57VNKK193 pKa = 10.5SS194 pKa = 3.03
Molecular weight: 21.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12572 |
62 |
3640 |
314.3 |
34.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.64 ± 0.46 | 0.628 ± 0.196 |
6.753 ± 0.28 | 7.564 ± 0.476 |
3.253 ± 0.214 | 7.652 ± 0.347 |
1.631 ± 0.208 | 5.234 ± 0.222 |
6.554 ± 0.364 | 7.222 ± 0.24 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.911 ± 0.168 | 4.184 ± 0.199 |
4.208 ± 0.22 | 5.393 ± 0.361 |
5.433 ± 0.402 | 5.409 ± 0.378 |
5.608 ± 0.282 | 6.244 ± 0.296 |
1.456 ± 0.193 | 3.023 ± 0.257 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
