
Cellulomonas sp. PhB150
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Cellulomonas; unclassified Cellulomonas
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
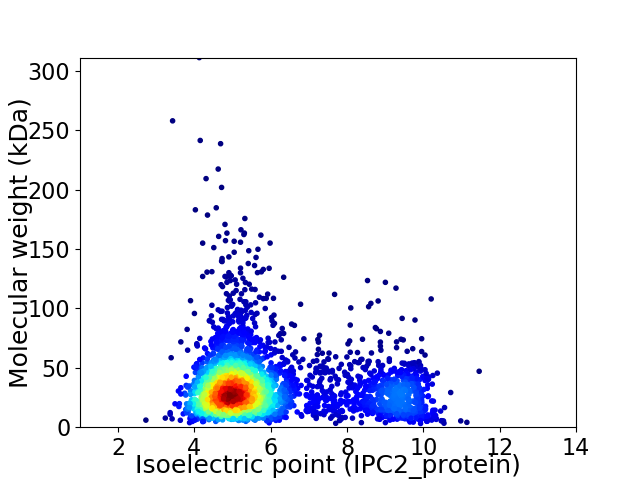
Virtual 2D-PAGE plot for 3418 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N2FI34|A0A3N2FI34_9CELL Enamine deaminase RidA (YjgF/YER057c/UK114 family) OS=Cellulomonas sp. PhB150 OX=2485188 GN=EDF34_3059 PE=4 SV=1
MM1 pKa = 7.7RR2 pKa = 11.84ALLVLVAASGLALAGCSSAPEE23 pKa = 4.29VVTSYY28 pKa = 11.73ADD30 pKa = 3.45YY31 pKa = 10.03PSYY34 pKa = 8.56EE35 pKa = 4.21TPQDD39 pKa = 3.18IVDD42 pKa = 3.81AADD45 pKa = 3.25VVVRR49 pKa = 11.84GEE51 pKa = 4.02VTGARR56 pKa = 11.84VKK58 pKa = 10.41EE59 pKa = 4.0DD60 pKa = 3.33QPIVSTTGDD69 pKa = 3.39PSLNPQAGLDD79 pKa = 3.74EE80 pKa = 4.78GTDD83 pKa = 3.55VPGVVVTISTVRR95 pKa = 11.84VEE97 pKa = 4.16EE98 pKa = 4.36VIKK101 pKa = 11.02GDD103 pKa = 3.66VAVGDD108 pKa = 4.28VIEE111 pKa = 4.2VAQLGGLYY119 pKa = 10.68DD120 pKa = 3.52GVRR123 pKa = 11.84YY124 pKa = 10.11VEE126 pKa = 4.32ASTVTLEE133 pKa = 4.52KK134 pKa = 11.04GSDD137 pKa = 3.49DD138 pKa = 4.3YY139 pKa = 11.67MLLLADD145 pKa = 4.96HH146 pKa = 6.97GPGVPYY152 pKa = 10.79DD153 pKa = 3.91LLHH156 pKa = 6.76PVQAMYY162 pKa = 9.79TVDD165 pKa = 5.06SGDD168 pKa = 4.26LDD170 pKa = 3.72PVADD174 pKa = 3.93NDD176 pKa = 4.03VAVDD180 pKa = 3.89TVDD183 pKa = 5.31DD184 pKa = 4.28LAEE187 pKa = 4.23LAQDD191 pKa = 3.44
MM1 pKa = 7.7RR2 pKa = 11.84ALLVLVAASGLALAGCSSAPEE23 pKa = 4.29VVTSYY28 pKa = 11.73ADD30 pKa = 3.45YY31 pKa = 10.03PSYY34 pKa = 8.56EE35 pKa = 4.21TPQDD39 pKa = 3.18IVDD42 pKa = 3.81AADD45 pKa = 3.25VVVRR49 pKa = 11.84GEE51 pKa = 4.02VTGARR56 pKa = 11.84VKK58 pKa = 10.41EE59 pKa = 4.0DD60 pKa = 3.33QPIVSTTGDD69 pKa = 3.39PSLNPQAGLDD79 pKa = 3.74EE80 pKa = 4.78GTDD83 pKa = 3.55VPGVVVTISTVRR95 pKa = 11.84VEE97 pKa = 4.16EE98 pKa = 4.36VIKK101 pKa = 11.02GDD103 pKa = 3.66VAVGDD108 pKa = 4.28VIEE111 pKa = 4.2VAQLGGLYY119 pKa = 10.68DD120 pKa = 3.52GVRR123 pKa = 11.84YY124 pKa = 10.11VEE126 pKa = 4.32ASTVTLEE133 pKa = 4.52KK134 pKa = 11.04GSDD137 pKa = 3.49DD138 pKa = 4.3YY139 pKa = 11.67MLLLADD145 pKa = 4.96HH146 pKa = 6.97GPGVPYY152 pKa = 10.79DD153 pKa = 3.91LLHH156 pKa = 6.76PVQAMYY162 pKa = 9.79TVDD165 pKa = 5.06SGDD168 pKa = 4.26LDD170 pKa = 3.72PVADD174 pKa = 3.93NDD176 pKa = 4.03VAVDD180 pKa = 3.89TVDD183 pKa = 5.31DD184 pKa = 4.28LAEE187 pKa = 4.23LAQDD191 pKa = 3.44
Molecular weight: 19.85 kDa
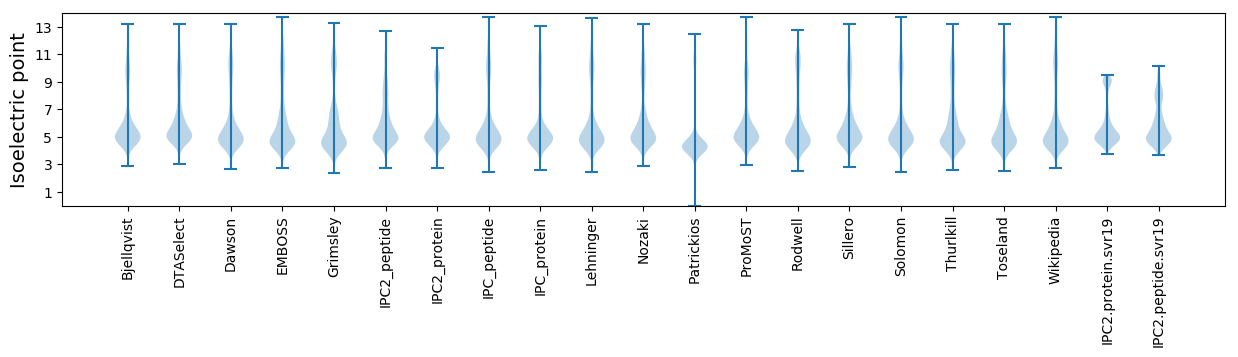
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N2G2V1|A0A3N2G2V1_9CELL Cysteine dioxygenase type I OS=Cellulomonas sp. PhB150 OX=2485188 GN=EDF34_0423 PE=3 SV=1
MM1 pKa = 7.08GRR3 pKa = 11.84AATVLVRR10 pKa = 11.84ATVPPMGRR18 pKa = 11.84AATVPARR25 pKa = 11.84ATVLPTATVSVMTAVAARR43 pKa = 11.84RR44 pKa = 11.84TGRR47 pKa = 11.84AATGHH52 pKa = 5.61GRR54 pKa = 11.84GIARR58 pKa = 11.84ATATVPAPVTVPLRR72 pKa = 11.84ADD74 pKa = 3.25VRR76 pKa = 11.84PTARR80 pKa = 11.84AVTVRR85 pKa = 11.84GPATVLPTVTVRR97 pKa = 11.84VTTAVAVRR105 pKa = 11.84PMARR109 pKa = 11.84AATARR114 pKa = 11.84ATTAVAVPRR123 pKa = 11.84TGRR126 pKa = 11.84ATARR130 pKa = 11.84AATGRR135 pKa = 11.84RR136 pKa = 11.84TATVPRR142 pKa = 11.84RR143 pKa = 11.84ATALARR149 pKa = 11.84AIVRR153 pKa = 11.84RR154 pKa = 11.84RR155 pKa = 11.84ATALARR161 pKa = 11.84AIVRR165 pKa = 11.84PTATVRR171 pKa = 11.84VTTVAAVPRR180 pKa = 11.84TGRR183 pKa = 11.84AATALARR190 pKa = 11.84VTVLPTAIDD199 pKa = 3.6RR200 pKa = 11.84ATTAVAARR208 pKa = 11.84PLGRR212 pKa = 11.84AATALVATARR222 pKa = 11.84PTATAPRR229 pKa = 11.84APRR232 pKa = 11.84RR233 pKa = 11.84ASVVTAPRR241 pKa = 11.84TATAVAVRR249 pKa = 11.84PTGRR253 pKa = 11.84ALIALGAAIVLPTVTARR270 pKa = 11.84AATGRR275 pKa = 11.84HH276 pKa = 3.9RR277 pKa = 11.84VAVRR281 pKa = 11.84RR282 pKa = 11.84TAIARR287 pKa = 11.84HH288 pKa = 4.64TAIARR293 pKa = 11.84HH294 pKa = 4.59TAIVRR299 pKa = 11.84RR300 pKa = 11.84PATARR305 pKa = 11.84HH306 pKa = 5.93APHH309 pKa = 7.09PVTVRR314 pKa = 11.84ARR316 pKa = 11.84TVRR319 pKa = 11.84PRR321 pKa = 11.84VTVRR325 pKa = 11.84RR326 pKa = 11.84TATARR331 pKa = 11.84ARR333 pKa = 11.84GTGARR338 pKa = 11.84SPASGRR344 pKa = 11.84ATVRR348 pKa = 11.84APRR351 pKa = 11.84ARR353 pKa = 11.84PPAASAPLGRR363 pKa = 11.84RR364 pKa = 11.84TARR367 pKa = 11.84LAAIVPRR374 pKa = 11.84TALVPRR380 pKa = 11.84RR381 pKa = 11.84AIDD384 pKa = 3.66PRR386 pKa = 11.84PVTAARR392 pKa = 11.84RR393 pKa = 11.84ATGMTGPTPVVATGRR408 pKa = 11.84IARR411 pKa = 11.84RR412 pKa = 11.84ARR414 pKa = 11.84TRR416 pKa = 11.84AAVRR420 pKa = 11.84ATVRR424 pKa = 11.84RR425 pKa = 11.84RR426 pKa = 11.84TDD428 pKa = 3.0RR429 pKa = 11.84SATAVRR435 pKa = 11.84PAATTTASPVRR446 pKa = 11.84RR447 pKa = 11.84RR448 pKa = 11.84TSGRR452 pKa = 11.84PRR454 pKa = 11.84RR455 pKa = 3.86
MM1 pKa = 7.08GRR3 pKa = 11.84AATVLVRR10 pKa = 11.84ATVPPMGRR18 pKa = 11.84AATVPARR25 pKa = 11.84ATVLPTATVSVMTAVAARR43 pKa = 11.84RR44 pKa = 11.84TGRR47 pKa = 11.84AATGHH52 pKa = 5.61GRR54 pKa = 11.84GIARR58 pKa = 11.84ATATVPAPVTVPLRR72 pKa = 11.84ADD74 pKa = 3.25VRR76 pKa = 11.84PTARR80 pKa = 11.84AVTVRR85 pKa = 11.84GPATVLPTVTVRR97 pKa = 11.84VTTAVAVRR105 pKa = 11.84PMARR109 pKa = 11.84AATARR114 pKa = 11.84ATTAVAVPRR123 pKa = 11.84TGRR126 pKa = 11.84ATARR130 pKa = 11.84AATGRR135 pKa = 11.84RR136 pKa = 11.84TATVPRR142 pKa = 11.84RR143 pKa = 11.84ATALARR149 pKa = 11.84AIVRR153 pKa = 11.84RR154 pKa = 11.84RR155 pKa = 11.84ATALARR161 pKa = 11.84AIVRR165 pKa = 11.84PTATVRR171 pKa = 11.84VTTVAAVPRR180 pKa = 11.84TGRR183 pKa = 11.84AATALARR190 pKa = 11.84VTVLPTAIDD199 pKa = 3.6RR200 pKa = 11.84ATTAVAARR208 pKa = 11.84PLGRR212 pKa = 11.84AATALVATARR222 pKa = 11.84PTATAPRR229 pKa = 11.84APRR232 pKa = 11.84RR233 pKa = 11.84ASVVTAPRR241 pKa = 11.84TATAVAVRR249 pKa = 11.84PTGRR253 pKa = 11.84ALIALGAAIVLPTVTARR270 pKa = 11.84AATGRR275 pKa = 11.84HH276 pKa = 3.9RR277 pKa = 11.84VAVRR281 pKa = 11.84RR282 pKa = 11.84TAIARR287 pKa = 11.84HH288 pKa = 4.64TAIARR293 pKa = 11.84HH294 pKa = 4.59TAIVRR299 pKa = 11.84RR300 pKa = 11.84PATARR305 pKa = 11.84HH306 pKa = 5.93APHH309 pKa = 7.09PVTVRR314 pKa = 11.84ARR316 pKa = 11.84TVRR319 pKa = 11.84PRR321 pKa = 11.84VTVRR325 pKa = 11.84RR326 pKa = 11.84TATARR331 pKa = 11.84ARR333 pKa = 11.84GTGARR338 pKa = 11.84SPASGRR344 pKa = 11.84ATVRR348 pKa = 11.84APRR351 pKa = 11.84ARR353 pKa = 11.84PPAASAPLGRR363 pKa = 11.84RR364 pKa = 11.84TARR367 pKa = 11.84LAAIVPRR374 pKa = 11.84TALVPRR380 pKa = 11.84RR381 pKa = 11.84AIDD384 pKa = 3.66PRR386 pKa = 11.84PVTAARR392 pKa = 11.84RR393 pKa = 11.84ATGMTGPTPVVATGRR408 pKa = 11.84IARR411 pKa = 11.84RR412 pKa = 11.84ARR414 pKa = 11.84TRR416 pKa = 11.84AAVRR420 pKa = 11.84ATVRR424 pKa = 11.84RR425 pKa = 11.84RR426 pKa = 11.84TDD428 pKa = 3.0RR429 pKa = 11.84SATAVRR435 pKa = 11.84PAATTTASPVRR446 pKa = 11.84RR447 pKa = 11.84RR448 pKa = 11.84TSGRR452 pKa = 11.84PRR454 pKa = 11.84RR455 pKa = 3.86
Molecular weight: 47.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1165880 |
29 |
3045 |
341.1 |
36.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.25 ± 0.065 | 0.535 ± 0.009 |
6.519 ± 0.041 | 5.007 ± 0.038 |
2.589 ± 0.024 | 9.232 ± 0.04 |
1.993 ± 0.022 | 3.321 ± 0.03 |
1.841 ± 0.036 | 10.071 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.544 ± 0.016 | 1.628 ± 0.028 |
5.728 ± 0.033 | 2.662 ± 0.021 |
7.204 ± 0.052 | 5.53 ± 0.04 |
6.759 ± 0.059 | 10.133 ± 0.041 |
1.58 ± 0.019 | 1.875 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
