
Burkholderia virus phiE202
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Peduovirinae; Tigrvirus
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
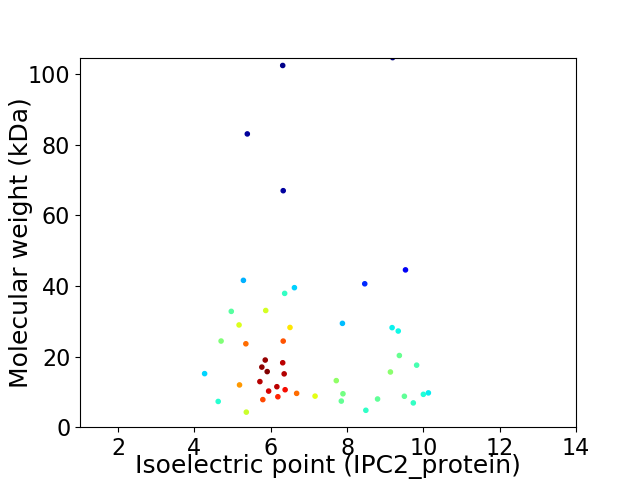
Virtual 2D-PAGE plot for 48 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A4JWT8|A4JWT8_9CAUD Gp41 LysC OS=Burkholderia virus phiE202 OX=431893 GN=BTHphiE202_0041 PE=4 SV=1
MM1 pKa = 7.55NKK3 pKa = 9.72PSSLRR8 pKa = 11.84AALVAALPQLNASPDD23 pKa = 3.54QLLVFVNEE31 pKa = 4.08GRR33 pKa = 11.84IEE35 pKa = 4.06ATGTRR40 pKa = 11.84TASFDD45 pKa = 3.57YY46 pKa = 10.55EE47 pKa = 4.45YY48 pKa = 10.09EE49 pKa = 4.1CEE51 pKa = 3.94IIIRR55 pKa = 11.84DD56 pKa = 4.52FIGNPDD62 pKa = 3.7DD63 pKa = 4.59VMIAVVEE70 pKa = 4.21WARR73 pKa = 11.84ANQPDD78 pKa = 4.44LVTNRR83 pKa = 11.84DD84 pKa = 3.08EE85 pKa = 4.6RR86 pKa = 11.84RR87 pKa = 11.84NGMTFVADD95 pKa = 3.33ILSNNAVDD103 pKa = 5.23LGLKK107 pKa = 9.89VKK109 pKa = 10.66LSEE112 pKa = 4.35SVVVGTDD119 pKa = 2.99EE120 pKa = 4.33AGNRR124 pKa = 11.84TVEE127 pKa = 4.78HH128 pKa = 6.93IDD130 pKa = 3.61DD131 pKa = 5.43AADD134 pKa = 3.14EE135 pKa = 4.32WLSS138 pKa = 3.44
MM1 pKa = 7.55NKK3 pKa = 9.72PSSLRR8 pKa = 11.84AALVAALPQLNASPDD23 pKa = 3.54QLLVFVNEE31 pKa = 4.08GRR33 pKa = 11.84IEE35 pKa = 4.06ATGTRR40 pKa = 11.84TASFDD45 pKa = 3.57YY46 pKa = 10.55EE47 pKa = 4.45YY48 pKa = 10.09EE49 pKa = 4.1CEE51 pKa = 3.94IIIRR55 pKa = 11.84DD56 pKa = 4.52FIGNPDD62 pKa = 3.7DD63 pKa = 4.59VMIAVVEE70 pKa = 4.21WARR73 pKa = 11.84ANQPDD78 pKa = 4.44LVTNRR83 pKa = 11.84DD84 pKa = 3.08EE85 pKa = 4.6RR86 pKa = 11.84RR87 pKa = 11.84NGMTFVADD95 pKa = 3.33ILSNNAVDD103 pKa = 5.23LGLKK107 pKa = 9.89VKK109 pKa = 10.66LSEE112 pKa = 4.35SVVVGTDD119 pKa = 2.99EE120 pKa = 4.33AGNRR124 pKa = 11.84TVEE127 pKa = 4.78HH128 pKa = 6.93IDD130 pKa = 3.61DD131 pKa = 5.43AADD134 pKa = 3.14EE135 pKa = 4.32WLSS138 pKa = 3.44
Molecular weight: 15.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A4JWQ8|A4JWQ8_9CAUD Toprim domain-containing protein OS=Burkholderia virus phiE202 OX=431893 GN=BTHphiE202_0011 PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84FYY4 pKa = 11.02KK5 pKa = 10.16RR6 pKa = 11.84QKK8 pKa = 9.85NGPWWFDD15 pKa = 3.05LSIDD19 pKa = 3.7GSRR22 pKa = 11.84IRR24 pKa = 11.84QSSGTSDD31 pKa = 2.75RR32 pKa = 11.84RR33 pKa = 11.84AAEE36 pKa = 3.71EE37 pKa = 3.76LAAKK41 pKa = 9.58VASDD45 pKa = 2.99YY46 pKa = 10.33WRR48 pKa = 11.84QKK50 pKa = 11.12KK51 pKa = 10.21LGEE54 pKa = 4.39RR55 pKa = 11.84PTVTWDD61 pKa = 3.01AAVVHH66 pKa = 6.13WLKK69 pKa = 10.89QNQHH73 pKa = 4.62QRR75 pKa = 11.84SLEE78 pKa = 4.1TTKK81 pKa = 10.4QRR83 pKa = 11.84LRR85 pKa = 11.84WLTDD89 pKa = 3.02QLKK92 pKa = 10.66GEE94 pKa = 4.22NVRR97 pKa = 11.84NIDD100 pKa = 3.52RR101 pKa = 11.84EE102 pKa = 4.15RR103 pKa = 11.84IQALIEE109 pKa = 3.81TKK111 pKa = 10.45AAEE114 pKa = 4.5KK115 pKa = 10.88YY116 pKa = 10.03RR117 pKa = 11.84GSPVAGATVNRR128 pKa = 11.84HH129 pKa = 4.03MAALSVILHH138 pKa = 6.28HH139 pKa = 6.37CHH141 pKa = 6.46AEE143 pKa = 3.93GWIDD147 pKa = 3.67AVPPIRR153 pKa = 11.84KK154 pKa = 8.83LRR156 pKa = 11.84EE157 pKa = 3.33NSARR161 pKa = 11.84LTWLTRR167 pKa = 11.84AQAQRR172 pKa = 11.84LLEE175 pKa = 4.15EE176 pKa = 4.95LPTHH180 pKa = 6.55LRR182 pKa = 11.84QMARR186 pKa = 11.84FALATGLRR194 pKa = 11.84EE195 pKa = 4.27SNVRR199 pKa = 11.84LLEE202 pKa = 3.9WAQVDD207 pKa = 4.41RR208 pKa = 11.84EE209 pKa = 4.03RR210 pKa = 11.84ALAWIHH216 pKa = 6.96ADD218 pKa = 3.13QAKK221 pKa = 9.28AGKK224 pKa = 9.85VISVPLNEE232 pKa = 4.97DD233 pKa = 2.97ALGVLRR239 pKa = 11.84EE240 pKa = 3.99QQGQHH245 pKa = 4.65KK246 pKa = 10.04RR247 pKa = 11.84YY248 pKa = 8.69VFVYY252 pKa = 9.12KK253 pKa = 10.36GAPIGRR259 pKa = 11.84IYY261 pKa = 10.26NHH263 pKa = 6.83AWQKK267 pKa = 9.5ACVRR271 pKa = 11.84AGLSGLRR278 pKa = 11.84FHH280 pKa = 7.58DD281 pKa = 5.35LRR283 pKa = 11.84HH284 pKa = 5.71TWASWHH290 pKa = 5.46VQAGTPLPILQQLGGWASYY309 pKa = 10.9QMVLRR314 pKa = 11.84YY315 pKa = 9.69AHH317 pKa = 7.51LGRR320 pKa = 11.84DD321 pKa = 3.49HH322 pKa = 6.31VAAYY326 pKa = 10.25ADD328 pKa = 3.74NIGTLRR334 pKa = 11.84HH335 pKa = 6.08KK336 pKa = 10.73SGTPPEE342 pKa = 4.1IEE344 pKa = 4.42KK345 pKa = 11.01GSDD348 pKa = 3.24CSEE351 pKa = 4.09PLSNLVAWGGIEE363 pKa = 3.84PPTRR367 pKa = 11.84GFSKK371 pKa = 9.85RR372 pKa = 11.84TADD375 pKa = 3.31TSAPSGKK382 pKa = 10.14ASRR385 pKa = 11.84PPKK388 pKa = 9.56KK389 pKa = 10.13RR390 pKa = 11.84KK391 pKa = 9.06AAA393 pKa = 3.76
MM1 pKa = 7.9RR2 pKa = 11.84FYY4 pKa = 11.02KK5 pKa = 10.16RR6 pKa = 11.84QKK8 pKa = 9.85NGPWWFDD15 pKa = 3.05LSIDD19 pKa = 3.7GSRR22 pKa = 11.84IRR24 pKa = 11.84QSSGTSDD31 pKa = 2.75RR32 pKa = 11.84RR33 pKa = 11.84AAEE36 pKa = 3.71EE37 pKa = 3.76LAAKK41 pKa = 9.58VASDD45 pKa = 2.99YY46 pKa = 10.33WRR48 pKa = 11.84QKK50 pKa = 11.12KK51 pKa = 10.21LGEE54 pKa = 4.39RR55 pKa = 11.84PTVTWDD61 pKa = 3.01AAVVHH66 pKa = 6.13WLKK69 pKa = 10.89QNQHH73 pKa = 4.62QRR75 pKa = 11.84SLEE78 pKa = 4.1TTKK81 pKa = 10.4QRR83 pKa = 11.84LRR85 pKa = 11.84WLTDD89 pKa = 3.02QLKK92 pKa = 10.66GEE94 pKa = 4.22NVRR97 pKa = 11.84NIDD100 pKa = 3.52RR101 pKa = 11.84EE102 pKa = 4.15RR103 pKa = 11.84IQALIEE109 pKa = 3.81TKK111 pKa = 10.45AAEE114 pKa = 4.5KK115 pKa = 10.88YY116 pKa = 10.03RR117 pKa = 11.84GSPVAGATVNRR128 pKa = 11.84HH129 pKa = 4.03MAALSVILHH138 pKa = 6.28HH139 pKa = 6.37CHH141 pKa = 6.46AEE143 pKa = 3.93GWIDD147 pKa = 3.67AVPPIRR153 pKa = 11.84KK154 pKa = 8.83LRR156 pKa = 11.84EE157 pKa = 3.33NSARR161 pKa = 11.84LTWLTRR167 pKa = 11.84AQAQRR172 pKa = 11.84LLEE175 pKa = 4.15EE176 pKa = 4.95LPTHH180 pKa = 6.55LRR182 pKa = 11.84QMARR186 pKa = 11.84FALATGLRR194 pKa = 11.84EE195 pKa = 4.27SNVRR199 pKa = 11.84LLEE202 pKa = 3.9WAQVDD207 pKa = 4.41RR208 pKa = 11.84EE209 pKa = 4.03RR210 pKa = 11.84ALAWIHH216 pKa = 6.96ADD218 pKa = 3.13QAKK221 pKa = 9.28AGKK224 pKa = 9.85VISVPLNEE232 pKa = 4.97DD233 pKa = 2.97ALGVLRR239 pKa = 11.84EE240 pKa = 3.99QQGQHH245 pKa = 4.65KK246 pKa = 10.04RR247 pKa = 11.84YY248 pKa = 8.69VFVYY252 pKa = 9.12KK253 pKa = 10.36GAPIGRR259 pKa = 11.84IYY261 pKa = 10.26NHH263 pKa = 6.83AWQKK267 pKa = 9.5ACVRR271 pKa = 11.84AGLSGLRR278 pKa = 11.84FHH280 pKa = 7.58DD281 pKa = 5.35LRR283 pKa = 11.84HH284 pKa = 5.71TWASWHH290 pKa = 5.46VQAGTPLPILQQLGGWASYY309 pKa = 10.9QMVLRR314 pKa = 11.84YY315 pKa = 9.69AHH317 pKa = 7.51LGRR320 pKa = 11.84DD321 pKa = 3.49HH322 pKa = 6.31VAAYY326 pKa = 10.25ADD328 pKa = 3.74NIGTLRR334 pKa = 11.84HH335 pKa = 6.08KK336 pKa = 10.73SGTPPEE342 pKa = 4.1IEE344 pKa = 4.42KK345 pKa = 11.01GSDD348 pKa = 3.24CSEE351 pKa = 4.09PLSNLVAWGGIEE363 pKa = 3.84PPTRR367 pKa = 11.84GFSKK371 pKa = 9.85RR372 pKa = 11.84TADD375 pKa = 3.31TSAPSGKK382 pKa = 10.14ASRR385 pKa = 11.84PPKK388 pKa = 9.56KK389 pKa = 10.13RR390 pKa = 11.84KK391 pKa = 9.06AAA393 pKa = 3.76
Molecular weight: 44.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10750 |
37 |
988 |
224.0 |
24.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.712 ± 0.565 | 0.828 ± 0.138 |
6.344 ± 0.336 | 5.321 ± 0.301 |
3.386 ± 0.242 | 7.656 ± 0.52 |
2.316 ± 0.214 | 4.019 ± 0.213 |
3.935 ± 0.264 | 8.837 ± 0.426 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.353 ± 0.249 | 2.986 ± 0.219 |
4.735 ± 0.293 | 3.414 ± 0.261 |
8.223 ± 0.363 | 5.051 ± 0.264 |
5.935 ± 0.269 | 6.893 ± 0.32 |
1.581 ± 0.168 | 2.474 ± 0.164 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
