
Manacus vitellinus (golden-collared manakin)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida; Sauria; Archelosauria; Archosauria; Dinosauria; Saurischia; Theropoda;
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
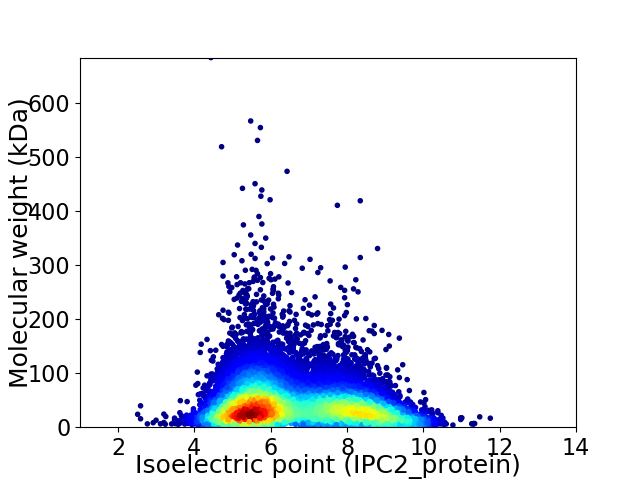
Virtual 2D-PAGE plot for 12707 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A093PW65|A0A093PW65_9PASS Nodal modulator 1 (Fragment) OS=Manacus vitellinus OX=328815 GN=N305_09670 PE=4 SV=1
CC1 pKa = 6.93NVGWAGNGNVCGPDD15 pKa = 3.43TDD17 pKa = 4.93LDD19 pKa = 4.37GYY21 pKa = 9.94PDD23 pKa = 4.1EE24 pKa = 5.31PLPCIDD30 pKa = 4.25NNKK33 pKa = 9.35HH34 pKa = 6.21CKK36 pKa = 9.14QDD38 pKa = 3.44NCRR41 pKa = 11.84LTPNSGQEE49 pKa = 3.95DD50 pKa = 4.26ADD52 pKa = 3.62NDD54 pKa = 5.11GIGDD58 pKa = 3.82QCDD61 pKa = 3.57DD62 pKa = 4.44DD63 pKa = 6.86ADD65 pKa = 4.16GDD67 pKa = 4.28GVKK70 pKa = 10.36NVEE73 pKa = 4.29DD74 pKa = 3.69NCRR77 pKa = 11.84LFPNKK82 pKa = 9.91DD83 pKa = 3.46QQNSDD88 pKa = 3.11TDD90 pKa = 4.03SFGDD94 pKa = 4.2ACDD97 pKa = 3.62NCPNVPNNDD106 pKa = 3.4QRR108 pKa = 11.84DD109 pKa = 3.62TDD111 pKa = 4.31SNGEE115 pKa = 3.91GDD117 pKa = 4.42ACDD120 pKa = 3.98NDD122 pKa = 3.39IDD124 pKa = 5.15GDD126 pKa = 4.46GIPNMLDD133 pKa = 3.17NCPKK137 pKa = 10.27VPNPLQTDD145 pKa = 3.4RR146 pKa = 11.84DD147 pKa = 3.67EE148 pKa = 6.42DD149 pKa = 4.4GVGDD153 pKa = 4.27ACDD156 pKa = 3.66SCPEE160 pKa = 4.12MSNPTQTDD168 pKa = 3.06MDD170 pKa = 4.69SDD172 pKa = 4.47LVGDD176 pKa = 3.68ICDD179 pKa = 3.85TNEE182 pKa = 4.14DD183 pKa = 3.74SDD185 pKa = 4.93GDD187 pKa = 3.91GHH189 pKa = 7.38QDD191 pKa = 3.18TKK193 pKa = 11.57DD194 pKa = 3.32NCAEE198 pKa = 4.25IPNSSQLDD206 pKa = 3.58SDD208 pKa = 4.01NDD210 pKa = 4.11GLGDD214 pKa = 4.98DD215 pKa = 5.48CDD217 pKa = 4.94NDD219 pKa = 4.37DD220 pKa = 5.42DD221 pKa = 5.52NDD223 pKa = 5.09GIPDD227 pKa = 4.02YY228 pKa = 11.2VPPGPDD234 pKa = 2.98NCRR237 pKa = 11.84LIPNPNQKK245 pKa = 10.66DD246 pKa = 3.36SDD248 pKa = 4.17GNGVGDD254 pKa = 3.6VCEE257 pKa = 4.36EE258 pKa = 4.28DD259 pKa = 3.68FDD261 pKa = 5.16NDD263 pKa = 4.06TVVDD267 pKa = 3.93QLDD270 pKa = 3.73VCPEE274 pKa = 3.98SAEE277 pKa = 4.05VTLTDD282 pKa = 3.82FRR284 pKa = 11.84AYY286 pKa = 8.36QTVILDD292 pKa = 3.91PEE294 pKa = 5.2GDD296 pKa = 3.69AQIDD300 pKa = 4.13PNWVVLNQGMEE311 pKa = 4.11IVQTMNSDD319 pKa = 3.19PGLAVGYY326 pKa = 7.31TAFNGVDD333 pKa = 3.99FEE335 pKa = 4.97GTFHH339 pKa = 6.81VNTVTDD345 pKa = 3.57DD346 pKa = 4.08DD347 pKa = 4.34YY348 pKa = 12.06AGFIFSYY355 pKa = 10.41QDD357 pKa = 2.92SASFYY362 pKa = 10.36VVMWKK367 pKa = 7.78QTEE370 pKa = 3.86QTYY373 pKa = 8.13WQATPFRR380 pKa = 11.84AVAEE384 pKa = 4.5PGLQLKK390 pKa = 9.47AVKK393 pKa = 10.34SSTGPGEE400 pKa = 4.21HH401 pKa = 6.64LRR403 pKa = 11.84NALWHH408 pKa = 6.26TGHH411 pKa = 6.51TPDD414 pKa = 5.16HH415 pKa = 6.71VRR417 pKa = 11.84LLWKK421 pKa = 10.36DD422 pKa = 3.2PRR424 pKa = 11.84NVGWRR429 pKa = 11.84DD430 pKa = 3.08KK431 pKa = 11.01TSYY434 pKa = 10.07RR435 pKa = 11.84WQLAHH440 pKa = 7.17RR441 pKa = 11.84PQVGYY446 pKa = 10.38IRR448 pKa = 4.61
CC1 pKa = 6.93NVGWAGNGNVCGPDD15 pKa = 3.43TDD17 pKa = 4.93LDD19 pKa = 4.37GYY21 pKa = 9.94PDD23 pKa = 4.1EE24 pKa = 5.31PLPCIDD30 pKa = 4.25NNKK33 pKa = 9.35HH34 pKa = 6.21CKK36 pKa = 9.14QDD38 pKa = 3.44NCRR41 pKa = 11.84LTPNSGQEE49 pKa = 3.95DD50 pKa = 4.26ADD52 pKa = 3.62NDD54 pKa = 5.11GIGDD58 pKa = 3.82QCDD61 pKa = 3.57DD62 pKa = 4.44DD63 pKa = 6.86ADD65 pKa = 4.16GDD67 pKa = 4.28GVKK70 pKa = 10.36NVEE73 pKa = 4.29DD74 pKa = 3.69NCRR77 pKa = 11.84LFPNKK82 pKa = 9.91DD83 pKa = 3.46QQNSDD88 pKa = 3.11TDD90 pKa = 4.03SFGDD94 pKa = 4.2ACDD97 pKa = 3.62NCPNVPNNDD106 pKa = 3.4QRR108 pKa = 11.84DD109 pKa = 3.62TDD111 pKa = 4.31SNGEE115 pKa = 3.91GDD117 pKa = 4.42ACDD120 pKa = 3.98NDD122 pKa = 3.39IDD124 pKa = 5.15GDD126 pKa = 4.46GIPNMLDD133 pKa = 3.17NCPKK137 pKa = 10.27VPNPLQTDD145 pKa = 3.4RR146 pKa = 11.84DD147 pKa = 3.67EE148 pKa = 6.42DD149 pKa = 4.4GVGDD153 pKa = 4.27ACDD156 pKa = 3.66SCPEE160 pKa = 4.12MSNPTQTDD168 pKa = 3.06MDD170 pKa = 4.69SDD172 pKa = 4.47LVGDD176 pKa = 3.68ICDD179 pKa = 3.85TNEE182 pKa = 4.14DD183 pKa = 3.74SDD185 pKa = 4.93GDD187 pKa = 3.91GHH189 pKa = 7.38QDD191 pKa = 3.18TKK193 pKa = 11.57DD194 pKa = 3.32NCAEE198 pKa = 4.25IPNSSQLDD206 pKa = 3.58SDD208 pKa = 4.01NDD210 pKa = 4.11GLGDD214 pKa = 4.98DD215 pKa = 5.48CDD217 pKa = 4.94NDD219 pKa = 4.37DD220 pKa = 5.42DD221 pKa = 5.52NDD223 pKa = 5.09GIPDD227 pKa = 4.02YY228 pKa = 11.2VPPGPDD234 pKa = 2.98NCRR237 pKa = 11.84LIPNPNQKK245 pKa = 10.66DD246 pKa = 3.36SDD248 pKa = 4.17GNGVGDD254 pKa = 3.6VCEE257 pKa = 4.36EE258 pKa = 4.28DD259 pKa = 3.68FDD261 pKa = 5.16NDD263 pKa = 4.06TVVDD267 pKa = 3.93QLDD270 pKa = 3.73VCPEE274 pKa = 3.98SAEE277 pKa = 4.05VTLTDD282 pKa = 3.82FRR284 pKa = 11.84AYY286 pKa = 8.36QTVILDD292 pKa = 3.91PEE294 pKa = 5.2GDD296 pKa = 3.69AQIDD300 pKa = 4.13PNWVVLNQGMEE311 pKa = 4.11IVQTMNSDD319 pKa = 3.19PGLAVGYY326 pKa = 7.31TAFNGVDD333 pKa = 3.99FEE335 pKa = 4.97GTFHH339 pKa = 6.81VNTVTDD345 pKa = 3.57DD346 pKa = 4.08DD347 pKa = 4.34YY348 pKa = 12.06AGFIFSYY355 pKa = 10.41QDD357 pKa = 2.92SASFYY362 pKa = 10.36VVMWKK367 pKa = 7.78QTEE370 pKa = 3.86QTYY373 pKa = 8.13WQATPFRR380 pKa = 11.84AVAEE384 pKa = 4.5PGLQLKK390 pKa = 9.47AVKK393 pKa = 10.34SSTGPGEE400 pKa = 4.21HH401 pKa = 6.64LRR403 pKa = 11.84NALWHH408 pKa = 6.26TGHH411 pKa = 6.51TPDD414 pKa = 5.16HH415 pKa = 6.71VRR417 pKa = 11.84LLWKK421 pKa = 10.36DD422 pKa = 3.2PRR424 pKa = 11.84NVGWRR429 pKa = 11.84DD430 pKa = 3.08KK431 pKa = 11.01TSYY434 pKa = 10.07RR435 pKa = 11.84WQLAHH440 pKa = 7.17RR441 pKa = 11.84PQVGYY446 pKa = 10.38IRR448 pKa = 4.61
Molecular weight: 49.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A093QBZ4|A0A093QBZ4_9PASS Zinc transporter 5 (Fragment) OS=Manacus vitellinus OX=328815 GN=N305_14703 PE=3 SV=1
GG1 pKa = 7.22KK2 pKa = 10.54GRR4 pKa = 11.84FGKK7 pKa = 10.68GRR9 pKa = 11.84FGKK12 pKa = 10.65GRR14 pKa = 11.84FGKK17 pKa = 10.65GRR19 pKa = 11.84FGKK22 pKa = 10.65GRR24 pKa = 11.84FGKK27 pKa = 10.65GRR29 pKa = 11.84FGKK32 pKa = 10.65GRR34 pKa = 11.84FGKK37 pKa = 10.65GRR39 pKa = 11.84FGKK42 pKa = 10.65GRR44 pKa = 11.84FGKK47 pKa = 10.65GRR49 pKa = 11.84FGKK52 pKa = 10.65GRR54 pKa = 11.84FGKK57 pKa = 10.7GRR59 pKa = 11.84FGG61 pKa = 3.6
GG1 pKa = 7.22KK2 pKa = 10.54GRR4 pKa = 11.84FGKK7 pKa = 10.68GRR9 pKa = 11.84FGKK12 pKa = 10.65GRR14 pKa = 11.84FGKK17 pKa = 10.65GRR19 pKa = 11.84FGKK22 pKa = 10.65GRR24 pKa = 11.84FGKK27 pKa = 10.65GRR29 pKa = 11.84FGKK32 pKa = 10.65GRR34 pKa = 11.84FGKK37 pKa = 10.65GRR39 pKa = 11.84FGKK42 pKa = 10.65GRR44 pKa = 11.84FGKK47 pKa = 10.65GRR49 pKa = 11.84FGKK52 pKa = 10.65GRR54 pKa = 11.84FGKK57 pKa = 10.7GRR59 pKa = 11.84FGG61 pKa = 3.6
Molecular weight: 6.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5377145 |
31 |
6219 |
423.2 |
47.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.43 ± 0.018 | 2.273 ± 0.021 |
5.026 ± 0.016 | 7.204 ± 0.03 |
3.92 ± 0.017 | 5.955 ± 0.027 |
2.57 ± 0.011 | 4.915 ± 0.018 |
6.367 ± 0.026 | 9.758 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.213 ± 0.008 | 4.058 ± 0.013 |
5.352 ± 0.024 | 4.676 ± 0.02 |
5.28 ± 0.018 | 8.144 ± 0.025 |
5.354 ± 0.015 | 6.325 ± 0.016 |
1.209 ± 0.008 | 2.968 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
