
Rheinheimera sp. SA_1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Chromatiaceae; Rheinheimera; unclassified Rheinheimera
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
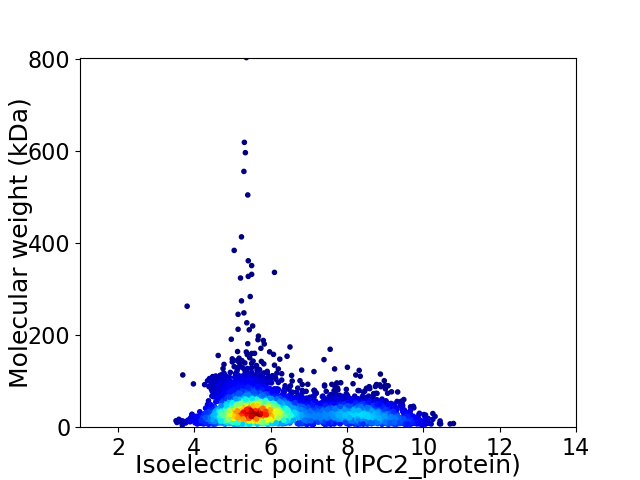
Virtual 2D-PAGE plot for 4186 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B7UHQ5|A0A1B7UHQ5_9GAMM Riboflavin biosynthesis protein OS=Rheinheimera sp. SA_1 OX=1827365 GN=A5320_15215 PE=3 SV=1
MM1 pKa = 7.25SLKK4 pKa = 10.28IAVFYY9 pKa = 10.99GSTTCYY15 pKa = 10.73TEE17 pKa = 3.71MTAEE21 pKa = 4.98KK22 pKa = 9.74IQALLAVDD30 pKa = 3.98AALPTGSSVTLHH42 pKa = 6.26NLKK45 pKa = 9.42QQPLATMANYY55 pKa = 10.13DD56 pKa = 3.79VLILGISTWDD66 pKa = 3.44FGEE69 pKa = 4.43LQEE72 pKa = 5.27DD73 pKa = 4.61WEE75 pKa = 4.39AHH77 pKa = 5.72WDD79 pKa = 4.03DD80 pKa = 3.82IAAVDD85 pKa = 4.03LSGKK89 pKa = 9.58IVAIYY94 pKa = 10.73GMGDD98 pKa = 3.17QLGYY102 pKa = 11.14AEE104 pKa = 4.8WFVDD108 pKa = 4.91AIGMLHH114 pKa = 5.81QAISDD119 pKa = 3.89QPCQRR124 pKa = 11.84IGFWSTQGYY133 pKa = 10.22DD134 pKa = 3.76FIKK137 pKa = 10.74SKK139 pKa = 11.09AVTDD143 pKa = 4.04DD144 pKa = 4.22GEE146 pKa = 4.34WFYY149 pKa = 11.91GLALDD154 pKa = 5.65EE155 pKa = 4.68EE156 pKa = 4.49NQYY159 pKa = 11.51DD160 pKa = 3.71QTDD163 pKa = 3.43EE164 pKa = 5.19RR165 pKa = 11.84LSLWLNQIVEE175 pKa = 4.24EE176 pKa = 4.25MIEE179 pKa = 4.11LVV181 pKa = 3.4
MM1 pKa = 7.25SLKK4 pKa = 10.28IAVFYY9 pKa = 10.99GSTTCYY15 pKa = 10.73TEE17 pKa = 3.71MTAEE21 pKa = 4.98KK22 pKa = 9.74IQALLAVDD30 pKa = 3.98AALPTGSSVTLHH42 pKa = 6.26NLKK45 pKa = 9.42QQPLATMANYY55 pKa = 10.13DD56 pKa = 3.79VLILGISTWDD66 pKa = 3.44FGEE69 pKa = 4.43LQEE72 pKa = 5.27DD73 pKa = 4.61WEE75 pKa = 4.39AHH77 pKa = 5.72WDD79 pKa = 4.03DD80 pKa = 3.82IAAVDD85 pKa = 4.03LSGKK89 pKa = 9.58IVAIYY94 pKa = 10.73GMGDD98 pKa = 3.17QLGYY102 pKa = 11.14AEE104 pKa = 4.8WFVDD108 pKa = 4.91AIGMLHH114 pKa = 5.81QAISDD119 pKa = 3.89QPCQRR124 pKa = 11.84IGFWSTQGYY133 pKa = 10.22DD134 pKa = 3.76FIKK137 pKa = 10.74SKK139 pKa = 11.09AVTDD143 pKa = 4.04DD144 pKa = 4.22GEE146 pKa = 4.34WFYY149 pKa = 11.91GLALDD154 pKa = 5.65EE155 pKa = 4.68EE156 pKa = 4.49NQYY159 pKa = 11.51DD160 pKa = 3.71QTDD163 pKa = 3.43EE164 pKa = 5.19RR165 pKa = 11.84LSLWLNQIVEE175 pKa = 4.24EE176 pKa = 4.25MIEE179 pKa = 4.11LVV181 pKa = 3.4
Molecular weight: 20.35 kDa
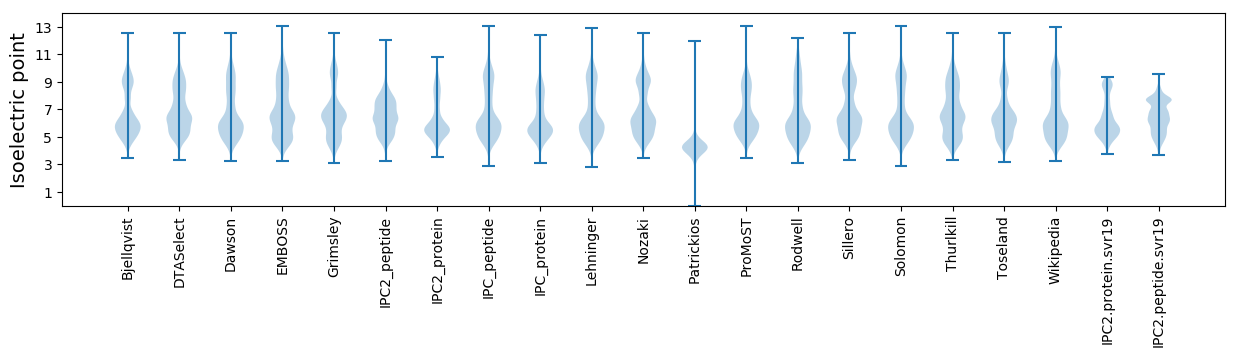
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B7UPC2|A0A1B7UPC2_9GAMM HlyD_D23 domain-containing protein OS=Rheinheimera sp. SA_1 OX=1827365 GN=A5320_02635 PE=4 SV=1
MM1 pKa = 7.7GSPFFCWRR9 pKa = 11.84LLALTLAPGTKK20 pKa = 9.66ARR22 pKa = 11.84QRR24 pKa = 11.84QIRR27 pKa = 11.84QRR29 pKa = 11.84LQSGNILLVKK39 pKa = 10.45RR40 pKa = 11.84SGPGKK45 pKa = 8.35TLLLPALVPARR56 pKa = 11.84WRR58 pKa = 11.84FSPVLFLMRR67 pKa = 11.84LQFF70 pKa = 4.12
MM1 pKa = 7.7GSPFFCWRR9 pKa = 11.84LLALTLAPGTKK20 pKa = 9.66ARR22 pKa = 11.84QRR24 pKa = 11.84QIRR27 pKa = 11.84QRR29 pKa = 11.84LQSGNILLVKK39 pKa = 10.45RR40 pKa = 11.84SGPGKK45 pKa = 8.35TLLLPALVPARR56 pKa = 11.84WRR58 pKa = 11.84FSPVLFLMRR67 pKa = 11.84LQFF70 pKa = 4.12
Molecular weight: 8.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1508662 |
37 |
7335 |
360.4 |
39.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.244 ± 0.042 | 0.994 ± 0.012 |
5.117 ± 0.031 | 5.029 ± 0.033 |
4.039 ± 0.026 | 6.653 ± 0.034 |
2.214 ± 0.019 | 5.175 ± 0.032 |
4.243 ± 0.041 | 11.996 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.242 ± 0.019 | 3.602 ± 0.028 |
4.432 ± 0.027 | 6.538 ± 0.06 |
4.719 ± 0.028 | 6.313 ± 0.03 |
5.257 ± 0.031 | 6.824 ± 0.03 |
1.435 ± 0.019 | 2.934 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
