
Methylobacterium haplocladii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Methylobacterium
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
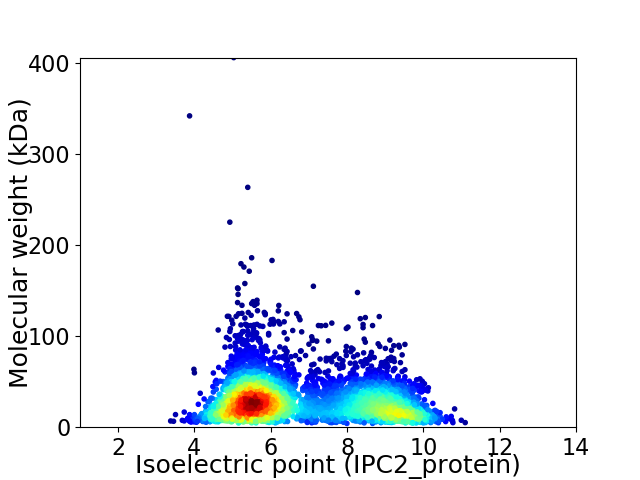
Virtual 2D-PAGE plot for 4347 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A512IVU1|A0A512IVU1_9RHIZ HcpA family protein OS=Methylobacterium haplocladii OX=1176176 GN=MHA02_41830 PE=4 SV=1
MM1 pKa = 7.06VLAGRR6 pKa = 11.84IWGTVMATAYY16 pKa = 7.34NTSVSDD22 pKa = 4.59LDD24 pKa = 3.54QHH26 pKa = 6.97PIDD29 pKa = 3.85PTTRR33 pKa = 11.84AEE35 pKa = 4.01IAQALALSQKK45 pKa = 9.04TGSDD49 pKa = 3.16TVTIEE54 pKa = 4.27APSTDD59 pKa = 2.95GTYY62 pKa = 10.35TLASNTNVFAATVSGSYY79 pKa = 9.21TLDD82 pKa = 3.21TSQSAGVPIIATGADD97 pKa = 3.65PVVLTLTGGVAADD110 pKa = 3.91VIVGSGGAYY119 pKa = 7.71VTGGTGNDD127 pKa = 3.95TIQGGVGSDD136 pKa = 3.43VLFSGYY142 pKa = 10.88AGGDD146 pKa = 3.39HH147 pKa = 6.99KK148 pKa = 10.94ISSGGGNDD156 pKa = 3.27SLYY159 pKa = 11.04GGTGADD165 pKa = 3.5TLTGGMGPGAGSNVLVGGTGKK186 pKa = 9.92NVLQGGDD193 pKa = 4.18GNDD196 pKa = 3.23SLYY199 pKa = 11.34GGGQSLLLAGSGKK212 pKa = 8.02QTLGGGYY219 pKa = 9.47YY220 pKa = 8.06QTAHH224 pKa = 6.58DD225 pKa = 4.17TLVGGTNNDD234 pKa = 3.28LLITFRR240 pKa = 11.84GNNTLSTGNGFGSDD254 pKa = 3.65TLIGGAGNDD263 pKa = 3.44TLTAVHH269 pKa = 6.6GNNVLFGGSTPDD281 pKa = 3.44ADD283 pKa = 3.88GKK285 pKa = 11.13LGISVLQGGDD295 pKa = 3.51GADD298 pKa = 3.14TLYY301 pKa = 11.32GGGRR305 pKa = 11.84SDD307 pKa = 4.88LRR309 pKa = 11.84AGSGNQTLGAGYY321 pKa = 9.53YY322 pKa = 11.0ANAQDD327 pKa = 4.12TLTGGSGNDD336 pKa = 3.84LLITFNGNNTLTTGSDD352 pKa = 3.49GSSDD356 pKa = 3.71TLIGGAGNDD365 pKa = 3.44TLTAVHH371 pKa = 6.63GNNVLVGGSGRR382 pKa = 11.84TLVFGGDD389 pKa = 3.64GNDD392 pKa = 3.34TLFGGGQSDD401 pKa = 3.95LRR403 pKa = 11.84AGTGNQTLGGGYY415 pKa = 9.07IDD417 pKa = 4.5NAHH420 pKa = 6.68DD421 pKa = 3.94TLTGGAGNDD430 pKa = 3.57LLITFKK436 pKa = 11.21GNNSLSTGTGSGHH449 pKa = 7.32DD450 pKa = 3.95SLFGGAGNDD459 pKa = 2.95TLTAVRR465 pKa = 11.84GDD467 pKa = 3.61NLLVAGSGKK476 pKa = 10.11SVLYY480 pKa = 10.78GGTGNDD486 pKa = 3.27TLYY489 pKa = 11.35GSGQSEE495 pKa = 4.02LHH497 pKa = 6.5AGSGSQTLGGGYY509 pKa = 9.64SAGAHH514 pKa = 5.52DD515 pKa = 4.34TLYY518 pKa = 11.03GGSGNSLLVVGQGNNQLTGGSGHH541 pKa = 7.41DD542 pKa = 3.58SLFGGSGNDD551 pKa = 3.55TLSAGANGSVLVAGTGNTLMQGSTGVDD578 pKa = 3.11QFGVSSLTGNDD589 pKa = 3.6TIVGGRR595 pKa = 11.84EE596 pKa = 3.89DD597 pKa = 3.31TLGIYY602 pKa = 10.13NHH604 pKa = 7.28DD605 pKa = 3.89SSAARR610 pKa = 11.84YY611 pKa = 5.74TQNSNGSTTITFEE624 pKa = 3.93GVANHH629 pKa = 6.86SITVSGLGSVTYY641 pKa = 11.02GDD643 pKa = 3.86GTTHH647 pKa = 7.56IFTGGNNSS655 pKa = 3.2
MM1 pKa = 7.06VLAGRR6 pKa = 11.84IWGTVMATAYY16 pKa = 7.34NTSVSDD22 pKa = 4.59LDD24 pKa = 3.54QHH26 pKa = 6.97PIDD29 pKa = 3.85PTTRR33 pKa = 11.84AEE35 pKa = 4.01IAQALALSQKK45 pKa = 9.04TGSDD49 pKa = 3.16TVTIEE54 pKa = 4.27APSTDD59 pKa = 2.95GTYY62 pKa = 10.35TLASNTNVFAATVSGSYY79 pKa = 9.21TLDD82 pKa = 3.21TSQSAGVPIIATGADD97 pKa = 3.65PVVLTLTGGVAADD110 pKa = 3.91VIVGSGGAYY119 pKa = 7.71VTGGTGNDD127 pKa = 3.95TIQGGVGSDD136 pKa = 3.43VLFSGYY142 pKa = 10.88AGGDD146 pKa = 3.39HH147 pKa = 6.99KK148 pKa = 10.94ISSGGGNDD156 pKa = 3.27SLYY159 pKa = 11.04GGTGADD165 pKa = 3.5TLTGGMGPGAGSNVLVGGTGKK186 pKa = 9.92NVLQGGDD193 pKa = 4.18GNDD196 pKa = 3.23SLYY199 pKa = 11.34GGGQSLLLAGSGKK212 pKa = 8.02QTLGGGYY219 pKa = 9.47YY220 pKa = 8.06QTAHH224 pKa = 6.58DD225 pKa = 4.17TLVGGTNNDD234 pKa = 3.28LLITFRR240 pKa = 11.84GNNTLSTGNGFGSDD254 pKa = 3.65TLIGGAGNDD263 pKa = 3.44TLTAVHH269 pKa = 6.6GNNVLFGGSTPDD281 pKa = 3.44ADD283 pKa = 3.88GKK285 pKa = 11.13LGISVLQGGDD295 pKa = 3.51GADD298 pKa = 3.14TLYY301 pKa = 11.32GGGRR305 pKa = 11.84SDD307 pKa = 4.88LRR309 pKa = 11.84AGSGNQTLGAGYY321 pKa = 9.53YY322 pKa = 11.0ANAQDD327 pKa = 4.12TLTGGSGNDD336 pKa = 3.84LLITFNGNNTLTTGSDD352 pKa = 3.49GSSDD356 pKa = 3.71TLIGGAGNDD365 pKa = 3.44TLTAVHH371 pKa = 6.63GNNVLVGGSGRR382 pKa = 11.84TLVFGGDD389 pKa = 3.64GNDD392 pKa = 3.34TLFGGGQSDD401 pKa = 3.95LRR403 pKa = 11.84AGTGNQTLGGGYY415 pKa = 9.07IDD417 pKa = 4.5NAHH420 pKa = 6.68DD421 pKa = 3.94TLTGGAGNDD430 pKa = 3.57LLITFKK436 pKa = 11.21GNNSLSTGTGSGHH449 pKa = 7.32DD450 pKa = 3.95SLFGGAGNDD459 pKa = 2.95TLTAVRR465 pKa = 11.84GDD467 pKa = 3.61NLLVAGSGKK476 pKa = 10.11SVLYY480 pKa = 10.78GGTGNDD486 pKa = 3.27TLYY489 pKa = 11.35GSGQSEE495 pKa = 4.02LHH497 pKa = 6.5AGSGSQTLGGGYY509 pKa = 9.64SAGAHH514 pKa = 5.52DD515 pKa = 4.34TLYY518 pKa = 11.03GGSGNSLLVVGQGNNQLTGGSGHH541 pKa = 7.41DD542 pKa = 3.58SLFGGSGNDD551 pKa = 3.55TLSAGANGSVLVAGTGNTLMQGSTGVDD578 pKa = 3.11QFGVSSLTGNDD589 pKa = 3.6TIVGGRR595 pKa = 11.84EE596 pKa = 3.89DD597 pKa = 3.31TLGIYY602 pKa = 10.13NHH604 pKa = 7.28DD605 pKa = 3.89SSAARR610 pKa = 11.84YY611 pKa = 5.74TQNSNGSTTITFEE624 pKa = 3.93GVANHH629 pKa = 6.86SITVSGLGSVTYY641 pKa = 11.02GDD643 pKa = 3.86GTTHH647 pKa = 7.56IFTGGNNSS655 pKa = 3.2
Molecular weight: 63.65 kDa
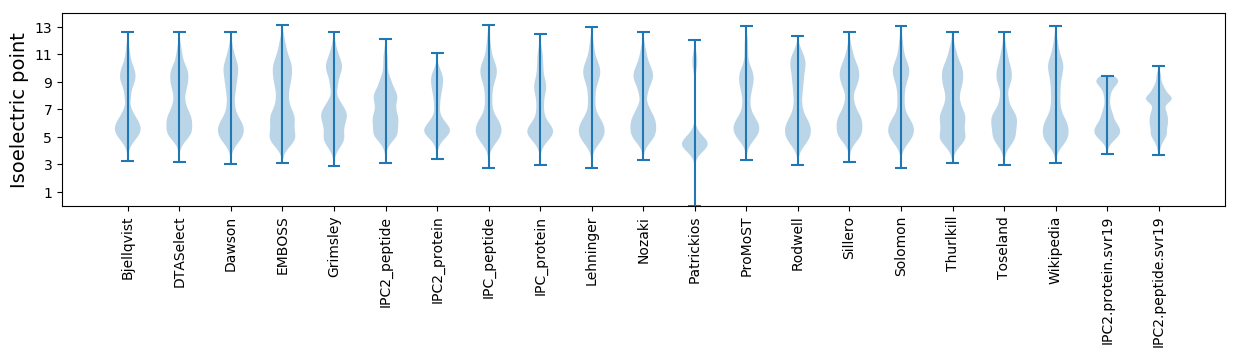
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A512IR11|A0A512IR11_9RHIZ Molybdenum cofactor sulfurase OS=Methylobacterium haplocladii OX=1176176 GN=MHA02_25420 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 9.15VIAARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.62GRR39 pKa = 11.84KK40 pKa = 9.35KK41 pKa = 10.66LSAA44 pKa = 3.99
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 9.15VIAARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.62GRR39 pKa = 11.84KK40 pKa = 9.35KK41 pKa = 10.66LSAA44 pKa = 3.99
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1297323 |
40 |
4283 |
298.4 |
32.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.543 ± 0.047 | 0.823 ± 0.012 |
5.744 ± 0.035 | 5.536 ± 0.04 |
3.463 ± 0.022 | 9.032 ± 0.057 |
1.914 ± 0.021 | 4.665 ± 0.023 |
2.908 ± 0.034 | 10.148 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.096 ± 0.017 | 2.213 ± 0.029 |
5.641 ± 0.038 | 2.782 ± 0.022 |
7.914 ± 0.046 | 5.299 ± 0.031 |
5.482 ± 0.052 | 7.537 ± 0.029 |
1.216 ± 0.015 | 2.044 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
