
Saitozyma podzolica
Taxonomy: cellular organisms; Eukaryota; <
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
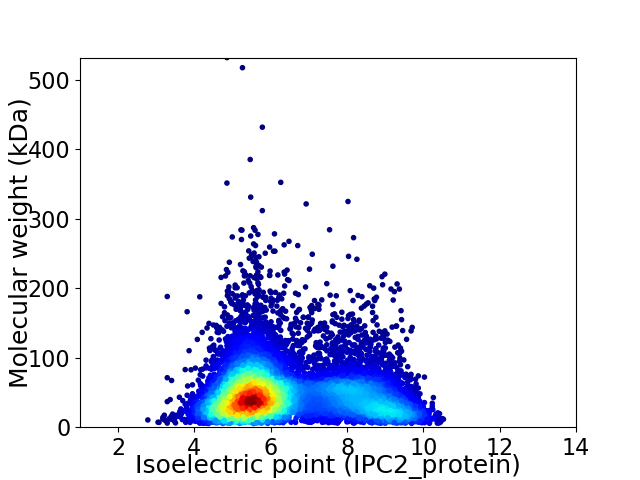
Virtual 2D-PAGE plot for 10205 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A427XL15|A0A427XL15_9TREE MFS domain-containing protein OS=Saitozyma podzolica OX=1890683 GN=EHS25_007968 PE=4 SV=1
MM1 pKa = 7.21ATVNSVSSMMSGMSSVSRR19 pKa = 11.84PLPMHH24 pKa = 6.47TAATPQPEE32 pKa = 4.39APTLGLGLDD41 pKa = 4.04LNLNPDD47 pKa = 3.34VGEE50 pKa = 4.45DD51 pKa = 3.28TDD53 pKa = 4.36GRR55 pKa = 11.84SDD57 pKa = 3.86PYY59 pKa = 10.36MGSPSSARR67 pKa = 11.84LQALNCSSSFLGFCMGSTITIPSTNTTLNPIFPEE101 pKa = 4.19LNPDD105 pKa = 2.87VDD107 pKa = 3.5IYY109 pKa = 7.95TARR112 pKa = 11.84MEE114 pKa = 4.36DD115 pKa = 3.01EE116 pKa = 4.44MMFSFSPQGVPGNSGDD132 pKa = 3.29VDD134 pKa = 3.61GFYY137 pKa = 10.34DD138 pKa = 4.04YY139 pKa = 11.24PFTPVMSTLPLPLPALGSLGPSQVNTANEE168 pKa = 4.49GTTMAATSQFLGDD181 pKa = 3.61SS182 pKa = 3.7
MM1 pKa = 7.21ATVNSVSSMMSGMSSVSRR19 pKa = 11.84PLPMHH24 pKa = 6.47TAATPQPEE32 pKa = 4.39APTLGLGLDD41 pKa = 4.04LNLNPDD47 pKa = 3.34VGEE50 pKa = 4.45DD51 pKa = 3.28TDD53 pKa = 4.36GRR55 pKa = 11.84SDD57 pKa = 3.86PYY59 pKa = 10.36MGSPSSARR67 pKa = 11.84LQALNCSSSFLGFCMGSTITIPSTNTTLNPIFPEE101 pKa = 4.19LNPDD105 pKa = 2.87VDD107 pKa = 3.5IYY109 pKa = 7.95TARR112 pKa = 11.84MEE114 pKa = 4.36DD115 pKa = 3.01EE116 pKa = 4.44MMFSFSPQGVPGNSGDD132 pKa = 3.29VDD134 pKa = 3.61GFYY137 pKa = 10.34DD138 pKa = 4.04YY139 pKa = 11.24PFTPVMSTLPLPLPALGSLGPSQVNTANEE168 pKa = 4.49GTTMAATSQFLGDD181 pKa = 3.61SS182 pKa = 3.7
Molecular weight: 19.04 kDa
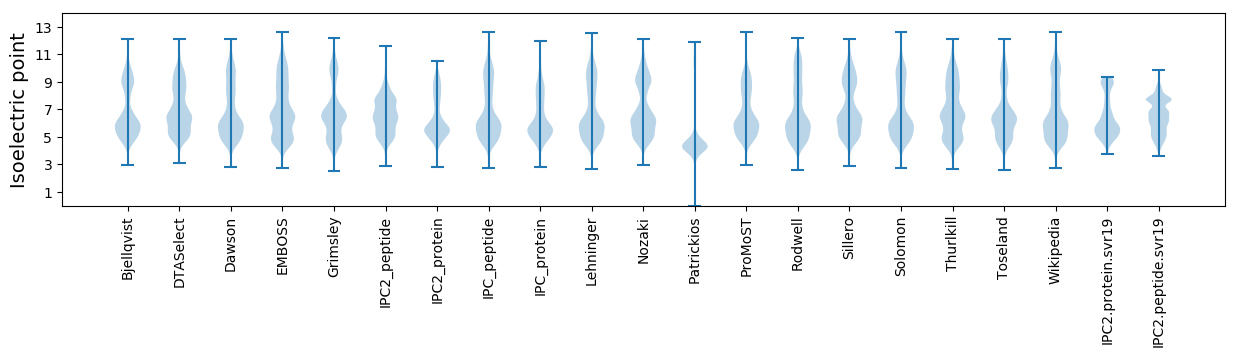
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A427Y3E9|A0A427Y3E9_9TREE Uncharacterized protein OS=Saitozyma podzolica OX=1890683 GN=EHS25_003738 PE=4 SV=1
MM1 pKa = 7.31SRR3 pKa = 11.84SQVGEE8 pKa = 3.77PLARR12 pKa = 11.84ASPIGTYY19 pKa = 10.47GPPCQSHH26 pKa = 6.36SLSDD30 pKa = 3.89QPPPLHH36 pKa = 6.78PPPHH40 pKa = 6.59LPPHH44 pKa = 6.29VGLSGVHH51 pKa = 5.54AHH53 pKa = 6.61HH54 pKa = 7.19PSSYY58 pKa = 8.88KK59 pKa = 9.71QSNGSVHH66 pKa = 6.86AGHH69 pKa = 7.23PHH71 pKa = 4.91THH73 pKa = 7.19ALPALPAPWRR83 pKa = 11.84STCRR87 pKa = 11.84TFPSPVAAGEE97 pKa = 4.04MSPTTFKK104 pKa = 11.22GGVAVKK110 pKa = 10.34RR111 pKa = 11.84GTAATKK117 pKa = 10.2GSQRR121 pKa = 11.84PARR124 pKa = 11.84DD125 pKa = 3.65HH126 pKa = 6.69EE127 pKa = 4.85SGSEE131 pKa = 3.95DD132 pKa = 3.55EE133 pKa = 4.63DD134 pKa = 3.88KK135 pKa = 11.06KK136 pKa = 11.45GKK138 pKa = 10.33SKK140 pKa = 11.29LEE142 pKa = 3.57NRR144 pKa = 11.84RR145 pKa = 11.84EE146 pKa = 4.07KK147 pKa = 11.16NRR149 pKa = 11.84VKK151 pKa = 10.3QRR153 pKa = 11.84KK154 pKa = 8.5LRR156 pKa = 11.84MRR158 pKa = 11.84RR159 pKa = 11.84ANHH162 pKa = 5.29IVEE165 pKa = 5.0LEE167 pKa = 3.98TSITSLRR174 pKa = 11.84SEE176 pKa = 4.2HH177 pKa = 6.49TSLQSAFSVSQLRR190 pKa = 11.84EE191 pKa = 3.54AHH193 pKa = 6.04LEE195 pKa = 3.83SRR197 pKa = 11.84IRR199 pKa = 11.84DD200 pKa = 3.38LGLALFSDD208 pKa = 4.68GLSGEE213 pKa = 4.06VDD215 pKa = 3.0ATRR218 pKa = 11.84RR219 pKa = 11.84AWSVVSSSSTPTGPLGCEE237 pKa = 4.19GGGKK241 pKa = 9.6RR242 pKa = 11.84PRR244 pKa = 11.84TISGRR249 pKa = 11.84DD250 pKa = 3.52PLNTLAQAALPLEE263 pKa = 4.58HH264 pKa = 6.86ARR266 pKa = 11.84EE267 pKa = 4.35GATLPPLVSQWNRR280 pKa = 11.84PSSRR284 pKa = 11.84HH285 pKa = 5.57SEE287 pKa = 3.98SDD289 pKa = 2.95EE290 pKa = 4.2KK291 pKa = 10.88KK292 pKa = 10.42CKK294 pKa = 10.2RR295 pKa = 11.84EE296 pKa = 3.95ALDD299 pKa = 3.75APLYY303 pKa = 10.16PPTFGKK309 pKa = 10.72SSTSRR314 pKa = 11.84GYY316 pKa = 10.71SRR318 pKa = 11.84TRR320 pKa = 11.84EE321 pKa = 3.81HH322 pKa = 6.38SPRR325 pKa = 11.84RR326 pKa = 11.84IPVEE330 pKa = 3.71PTTAGASSAGISEE343 pKa = 4.37PLVNPSLRR351 pKa = 11.84SSRR354 pKa = 11.84VASAPLPPLAPSSSSNSLFTTPEE377 pKa = 3.39WAAPSPGFIRR387 pKa = 11.84IEE389 pKa = 4.18DD390 pKa = 4.3LLSLMGPAAASLRR403 pKa = 11.84SQAHH407 pKa = 4.63TPKK410 pKa = 10.51KK411 pKa = 9.54RR412 pKa = 11.84DD413 pKa = 3.33QEE415 pKa = 4.17RR416 pKa = 11.84EE417 pKa = 3.76RR418 pKa = 11.84EE419 pKa = 4.54GISDD423 pKa = 3.56EE424 pKa = 4.04AVLRR428 pKa = 11.84RR429 pKa = 11.84RR430 pKa = 11.84GFGGRR435 pKa = 11.84PVPRR439 pKa = 11.84GRR441 pKa = 11.84PRR443 pKa = 11.84EE444 pKa = 4.25SRR446 pKa = 11.84CHH448 pKa = 6.04ASSCGRR454 pKa = 11.84CRR456 pKa = 11.84PRR458 pKa = 11.84FAVPVIIPPPDD469 pKa = 3.8PPVSLLLCPHH479 pKa = 6.96RR480 pKa = 11.84PLAPAALVPFSLAIPFATPLAPDD503 pKa = 3.8TANHH507 pKa = 6.25SKK509 pKa = 10.57LALSRR514 pKa = 11.84CPTSPHH520 pKa = 6.44HH521 pKa = 6.52LLPSTTRR528 pKa = 11.84PAAPQPLTRR537 pKa = 11.84RR538 pKa = 11.84RR539 pKa = 11.84KK540 pKa = 4.89THH542 pKa = 5.52RR543 pKa = 11.84RR544 pKa = 11.84RR545 pKa = 11.84PILLFRR551 pKa = 11.84RR552 pKa = 11.84RR553 pKa = 11.84ASRR556 pKa = 11.84ALSHH560 pKa = 6.31HH561 pKa = 6.45HH562 pKa = 6.89PAGQGHH568 pKa = 7.16PAPPQRR574 pKa = 11.84NADD577 pKa = 3.54SCLEE581 pKa = 4.69LISHH585 pKa = 6.71TLASVRR591 pKa = 11.84EE592 pKa = 4.42SGDD595 pKa = 3.49PPDD598 pKa = 4.26AEE600 pKa = 3.97AWEE603 pKa = 4.47LQDD606 pKa = 5.55SYY608 pKa = 11.24WDD610 pKa = 3.33TARR613 pKa = 11.84ASFRR617 pKa = 11.84IPASHH622 pKa = 6.88HH623 pKa = 5.92SFSILSQSEE632 pKa = 3.91HH633 pKa = 6.21AGARR637 pKa = 11.84LHH639 pKa = 6.55ALSSALFSRR648 pKa = 11.84VPRR651 pKa = 11.84TNLTRR656 pKa = 11.84SPFVDD661 pKa = 3.62LRR663 pKa = 11.84RR664 pKa = 11.84RR665 pKa = 11.84LSLRR669 pKa = 11.84YY670 pKa = 8.95IAIGSRR676 pKa = 11.84PRR678 pKa = 11.84HH679 pKa = 4.7RR680 pKa = 11.84TEE682 pKa = 4.76ALASTPGTRR691 pKa = 11.84VRR693 pKa = 11.84LEE695 pKa = 3.99PLGHH699 pKa = 4.73QTSRR703 pKa = 11.84RR704 pKa = 11.84NDD706 pKa = 3.11IQVITLLGSQATGLANAEE724 pKa = 3.85YY725 pKa = 10.46DD726 pKa = 3.76LTVVSLASKK735 pKa = 9.49DD736 pKa = 3.59ACSMSLPDD744 pKa = 4.52QDD746 pKa = 4.75GNPPRR751 pKa = 11.84LVNKK755 pKa = 10.21YY756 pKa = 9.06LDD758 pKa = 3.63SVADD762 pKa = 3.41HH763 pKa = 5.78TTRR766 pKa = 11.84SGTAQPAISRR776 pKa = 11.84STHH779 pKa = 5.61CLLPRR784 pKa = 11.84RR785 pKa = 11.84HH786 pKa = 7.07DD787 pKa = 3.62EE788 pKa = 4.27RR789 pKa = 11.84QHH791 pKa = 5.99HH792 pKa = 6.36QGLCLLEE799 pKa = 4.45TGNDD803 pKa = 3.14TWNIQPDD810 pKa = 4.12GQTVEE815 pKa = 4.3PLPFAGEE822 pKa = 4.68GPQLL826 pKa = 3.61
MM1 pKa = 7.31SRR3 pKa = 11.84SQVGEE8 pKa = 3.77PLARR12 pKa = 11.84ASPIGTYY19 pKa = 10.47GPPCQSHH26 pKa = 6.36SLSDD30 pKa = 3.89QPPPLHH36 pKa = 6.78PPPHH40 pKa = 6.59LPPHH44 pKa = 6.29VGLSGVHH51 pKa = 5.54AHH53 pKa = 6.61HH54 pKa = 7.19PSSYY58 pKa = 8.88KK59 pKa = 9.71QSNGSVHH66 pKa = 6.86AGHH69 pKa = 7.23PHH71 pKa = 4.91THH73 pKa = 7.19ALPALPAPWRR83 pKa = 11.84STCRR87 pKa = 11.84TFPSPVAAGEE97 pKa = 4.04MSPTTFKK104 pKa = 11.22GGVAVKK110 pKa = 10.34RR111 pKa = 11.84GTAATKK117 pKa = 10.2GSQRR121 pKa = 11.84PARR124 pKa = 11.84DD125 pKa = 3.65HH126 pKa = 6.69EE127 pKa = 4.85SGSEE131 pKa = 3.95DD132 pKa = 3.55EE133 pKa = 4.63DD134 pKa = 3.88KK135 pKa = 11.06KK136 pKa = 11.45GKK138 pKa = 10.33SKK140 pKa = 11.29LEE142 pKa = 3.57NRR144 pKa = 11.84RR145 pKa = 11.84EE146 pKa = 4.07KK147 pKa = 11.16NRR149 pKa = 11.84VKK151 pKa = 10.3QRR153 pKa = 11.84KK154 pKa = 8.5LRR156 pKa = 11.84MRR158 pKa = 11.84RR159 pKa = 11.84ANHH162 pKa = 5.29IVEE165 pKa = 5.0LEE167 pKa = 3.98TSITSLRR174 pKa = 11.84SEE176 pKa = 4.2HH177 pKa = 6.49TSLQSAFSVSQLRR190 pKa = 11.84EE191 pKa = 3.54AHH193 pKa = 6.04LEE195 pKa = 3.83SRR197 pKa = 11.84IRR199 pKa = 11.84DD200 pKa = 3.38LGLALFSDD208 pKa = 4.68GLSGEE213 pKa = 4.06VDD215 pKa = 3.0ATRR218 pKa = 11.84RR219 pKa = 11.84AWSVVSSSSTPTGPLGCEE237 pKa = 4.19GGGKK241 pKa = 9.6RR242 pKa = 11.84PRR244 pKa = 11.84TISGRR249 pKa = 11.84DD250 pKa = 3.52PLNTLAQAALPLEE263 pKa = 4.58HH264 pKa = 6.86ARR266 pKa = 11.84EE267 pKa = 4.35GATLPPLVSQWNRR280 pKa = 11.84PSSRR284 pKa = 11.84HH285 pKa = 5.57SEE287 pKa = 3.98SDD289 pKa = 2.95EE290 pKa = 4.2KK291 pKa = 10.88KK292 pKa = 10.42CKK294 pKa = 10.2RR295 pKa = 11.84EE296 pKa = 3.95ALDD299 pKa = 3.75APLYY303 pKa = 10.16PPTFGKK309 pKa = 10.72SSTSRR314 pKa = 11.84GYY316 pKa = 10.71SRR318 pKa = 11.84TRR320 pKa = 11.84EE321 pKa = 3.81HH322 pKa = 6.38SPRR325 pKa = 11.84RR326 pKa = 11.84IPVEE330 pKa = 3.71PTTAGASSAGISEE343 pKa = 4.37PLVNPSLRR351 pKa = 11.84SSRR354 pKa = 11.84VASAPLPPLAPSSSSNSLFTTPEE377 pKa = 3.39WAAPSPGFIRR387 pKa = 11.84IEE389 pKa = 4.18DD390 pKa = 4.3LLSLMGPAAASLRR403 pKa = 11.84SQAHH407 pKa = 4.63TPKK410 pKa = 10.51KK411 pKa = 9.54RR412 pKa = 11.84DD413 pKa = 3.33QEE415 pKa = 4.17RR416 pKa = 11.84EE417 pKa = 3.76RR418 pKa = 11.84EE419 pKa = 4.54GISDD423 pKa = 3.56EE424 pKa = 4.04AVLRR428 pKa = 11.84RR429 pKa = 11.84RR430 pKa = 11.84GFGGRR435 pKa = 11.84PVPRR439 pKa = 11.84GRR441 pKa = 11.84PRR443 pKa = 11.84EE444 pKa = 4.25SRR446 pKa = 11.84CHH448 pKa = 6.04ASSCGRR454 pKa = 11.84CRR456 pKa = 11.84PRR458 pKa = 11.84FAVPVIIPPPDD469 pKa = 3.8PPVSLLLCPHH479 pKa = 6.96RR480 pKa = 11.84PLAPAALVPFSLAIPFATPLAPDD503 pKa = 3.8TANHH507 pKa = 6.25SKK509 pKa = 10.57LALSRR514 pKa = 11.84CPTSPHH520 pKa = 6.44HH521 pKa = 6.52LLPSTTRR528 pKa = 11.84PAAPQPLTRR537 pKa = 11.84RR538 pKa = 11.84RR539 pKa = 11.84KK540 pKa = 4.89THH542 pKa = 5.52RR543 pKa = 11.84RR544 pKa = 11.84RR545 pKa = 11.84PILLFRR551 pKa = 11.84RR552 pKa = 11.84RR553 pKa = 11.84ASRR556 pKa = 11.84ALSHH560 pKa = 6.31HH561 pKa = 6.45HH562 pKa = 6.89PAGQGHH568 pKa = 7.16PAPPQRR574 pKa = 11.84NADD577 pKa = 3.54SCLEE581 pKa = 4.69LISHH585 pKa = 6.71TLASVRR591 pKa = 11.84EE592 pKa = 4.42SGDD595 pKa = 3.49PPDD598 pKa = 4.26AEE600 pKa = 3.97AWEE603 pKa = 4.47LQDD606 pKa = 5.55SYY608 pKa = 11.24WDD610 pKa = 3.33TARR613 pKa = 11.84ASFRR617 pKa = 11.84IPASHH622 pKa = 6.88HH623 pKa = 5.92SFSILSQSEE632 pKa = 3.91HH633 pKa = 6.21AGARR637 pKa = 11.84LHH639 pKa = 6.55ALSSALFSRR648 pKa = 11.84VPRR651 pKa = 11.84TNLTRR656 pKa = 11.84SPFVDD661 pKa = 3.62LRR663 pKa = 11.84RR664 pKa = 11.84RR665 pKa = 11.84LSLRR669 pKa = 11.84YY670 pKa = 8.95IAIGSRR676 pKa = 11.84PRR678 pKa = 11.84HH679 pKa = 4.7RR680 pKa = 11.84TEE682 pKa = 4.76ALASTPGTRR691 pKa = 11.84VRR693 pKa = 11.84LEE695 pKa = 3.99PLGHH699 pKa = 4.73QTSRR703 pKa = 11.84RR704 pKa = 11.84NDD706 pKa = 3.11IQVITLLGSQATGLANAEE724 pKa = 3.85YY725 pKa = 10.46DD726 pKa = 3.76LTVVSLASKK735 pKa = 9.49DD736 pKa = 3.59ACSMSLPDD744 pKa = 4.52QDD746 pKa = 4.75GNPPRR751 pKa = 11.84LVNKK755 pKa = 10.21YY756 pKa = 9.06LDD758 pKa = 3.63SVADD762 pKa = 3.41HH763 pKa = 5.78TTRR766 pKa = 11.84SGTAQPAISRR776 pKa = 11.84STHH779 pKa = 5.61CLLPRR784 pKa = 11.84RR785 pKa = 11.84HH786 pKa = 7.07DD787 pKa = 3.62EE788 pKa = 4.27RR789 pKa = 11.84QHH791 pKa = 5.99HH792 pKa = 6.36QGLCLLEE799 pKa = 4.45TGNDD803 pKa = 3.14TWNIQPDD810 pKa = 4.12GQTVEE815 pKa = 4.3PLPFAGEE822 pKa = 4.68GPQLL826 pKa = 3.61
Molecular weight: 89.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5014707 |
50 |
4841 |
491.4 |
53.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.624 ± 0.026 | 1.044 ± 0.008 |
5.508 ± 0.018 | 6.08 ± 0.022 |
3.34 ± 0.015 | 7.84 ± 0.021 |
2.393 ± 0.011 | 4.247 ± 0.015 |
4.057 ± 0.02 | 8.982 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.114 ± 0.01 | 2.879 ± 0.011 |
6.95 ± 0.034 | 3.535 ± 0.019 |
6.442 ± 0.022 | 8.728 ± 0.031 |
5.958 ± 0.017 | 6.415 ± 0.016 |
1.487 ± 0.01 | 2.376 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
