
Sodalis phage phiSG1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 51 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
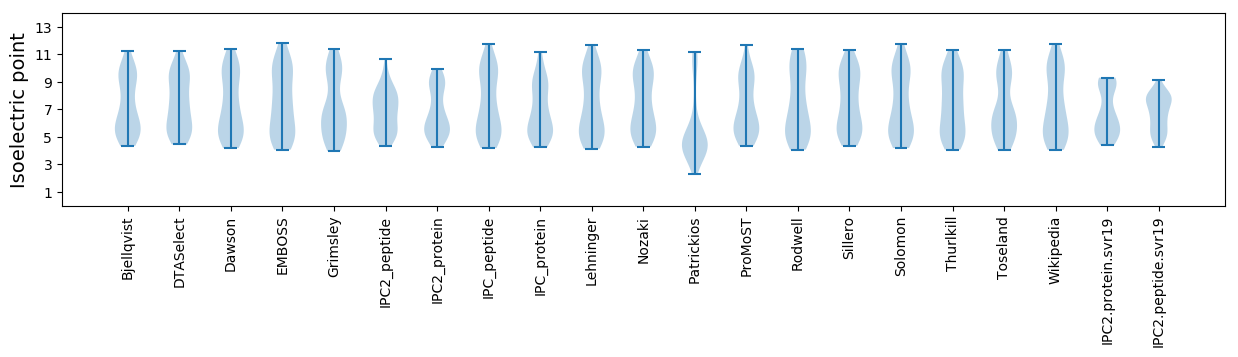
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A3E2I5|A3E2I5_9CAUD Gp23 OS=Sodalis phage phiSG1 OX=373126 PE=4 SV=1
MM1 pKa = 7.72PKK3 pKa = 10.27DD4 pKa = 3.49ISLDD8 pKa = 4.05DD9 pKa = 3.59IKK11 pKa = 11.1EE12 pKa = 3.95IALRR16 pKa = 11.84AYY18 pKa = 10.17RR19 pKa = 11.84GGEE22 pKa = 3.84VRR24 pKa = 11.84SAYY27 pKa = 9.99VDD29 pKa = 3.46KK30 pKa = 11.45SDD32 pKa = 4.22DD33 pKa = 3.4NSLISEE39 pKa = 4.84EE40 pKa = 4.18KK41 pKa = 10.06TEE43 pKa = 4.38LPPEE47 pKa = 4.41SVDD50 pKa = 3.68NPEE53 pKa = 3.86EE54 pKa = 3.91NAPEE58 pKa = 4.03SEE60 pKa = 4.33EE61 pKa = 3.98TVEE64 pKa = 4.08AVEE67 pKa = 5.09DD68 pKa = 3.8ATTVTDD74 pKa = 5.63SPDD77 pKa = 3.76CDD79 pKa = 3.86TQSSEE84 pKa = 4.6SEE86 pKa = 3.94EE87 pKa = 4.12TKK89 pKa = 11.05ARR91 pKa = 11.84DD92 pKa = 3.54WPDD95 pKa = 3.27LEE97 pKa = 5.78LDD99 pKa = 3.12IARR102 pKa = 11.84ALWEE106 pKa = 4.98GEE108 pKa = 4.23LHH110 pKa = 6.23WEE112 pKa = 5.11FIPSEE117 pKa = 4.15VEE119 pKa = 3.26QWARR123 pKa = 11.84KK124 pKa = 6.06MVKK127 pKa = 9.99RR128 pKa = 11.84RR129 pKa = 11.84DD130 pKa = 3.53GEE132 pKa = 4.07MMRR135 pKa = 11.84WLLVLQQVPDD145 pKa = 3.17ILSRR149 pKa = 11.84SRR151 pKa = 11.84EE152 pKa = 3.99FIFKK156 pKa = 10.1VIRR159 pKa = 11.84SPHH162 pKa = 6.26DD163 pKa = 3.26PEE165 pKa = 6.47IYY167 pKa = 8.87LTPEE171 pKa = 4.66AIDD174 pKa = 4.22AYY176 pKa = 9.96VASFVEE182 pKa = 4.2QEE184 pKa = 4.41TVEE187 pKa = 4.07EE188 pKa = 4.61VPWSEE193 pKa = 4.43EE194 pKa = 3.49ATADD198 pKa = 3.28IAEE201 pKa = 4.37TLTLEE206 pKa = 4.8RR207 pKa = 11.84DD208 pKa = 3.57AQRR211 pKa = 11.84LAKK214 pKa = 10.04FLEE217 pKa = 4.68PEE219 pKa = 4.17AVDD222 pKa = 4.66LPEE225 pKa = 6.35GPDD228 pKa = 4.9ADD230 pKa = 4.86DD231 pKa = 3.36EE232 pKa = 4.55WVPGVPEE239 pKa = 4.68GEE241 pKa = 4.34PEE243 pKa = 4.14TTVEE247 pKa = 4.91SEE249 pKa = 4.07TADD252 pKa = 3.47APDD255 pKa = 3.17IAEE258 pKa = 4.67PEE260 pKa = 4.42TVDD263 pKa = 4.52AEE265 pKa = 4.28ATEE268 pKa = 4.25SEE270 pKa = 4.65IYY272 pKa = 10.52YY273 pKa = 10.55SPEE276 pKa = 3.25TDD278 pKa = 2.95MPTFLRR284 pKa = 11.84DD285 pKa = 3.41LTPVNGAEE293 pKa = 3.81EE294 pKa = 4.45DD295 pKa = 3.59GRR297 pKa = 11.84LFATVDD303 pKa = 3.39ALNEE307 pKa = 3.97RR308 pKa = 11.84LAGIPAGGSLVLWGLDD324 pKa = 2.87NGLNHH329 pKa = 6.57SADD332 pKa = 4.5GYY334 pKa = 10.77SSTQIRR340 pKa = 11.84IALRR344 pKa = 11.84RR345 pKa = 11.84GKK347 pKa = 9.32PAATYY352 pKa = 8.87WYY354 pKa = 9.2KK355 pKa = 10.71DD356 pKa = 3.12APRR359 pKa = 11.84RR360 pKa = 11.84QEE362 pKa = 3.58DD363 pKa = 3.3SRR365 pKa = 11.84ALFIGSAVHH374 pKa = 6.81AALLEE379 pKa = 4.1PEE381 pKa = 4.47RR382 pKa = 11.84FATEE386 pKa = 4.02YY387 pKa = 11.31ACAPSVNLRR396 pKa = 11.84TSEE399 pKa = 4.01GKK401 pKa = 8.05EE402 pKa = 3.66TLASFEE408 pKa = 4.51CDD410 pKa = 2.88CATRR414 pKa = 11.84NLIPIKK420 pKa = 10.58RR421 pKa = 11.84EE422 pKa = 3.97DD423 pKa = 3.9FDD425 pKa = 4.7TVCMMRR431 pKa = 11.84DD432 pKa = 3.26SVLAFPVVRR441 pKa = 11.84LLLSSGVGEE450 pKa = 4.72LSVFWRR456 pKa = 11.84MEE458 pKa = 3.64NGLLLKK464 pKa = 10.12IRR466 pKa = 11.84PDD468 pKa = 3.1WLGVYY473 pKa = 9.91QDD475 pKa = 4.44APFMLDD481 pKa = 3.52VKK483 pKa = 10.28TLDD486 pKa = 3.75DD487 pKa = 3.88VNDD490 pKa = 5.01FGRR493 pKa = 11.84QAHH496 pKa = 4.86EE497 pKa = 4.08HH498 pKa = 6.64GYY500 pKa = 10.09HH501 pKa = 5.8IQAAYY506 pKa = 9.87YY507 pKa = 10.23GFVLEE512 pKa = 5.15HH513 pKa = 5.99VFGMVINGLFCTIGKK528 pKa = 8.5RR529 pKa = 11.84TEE531 pKa = 3.86CGRR534 pKa = 11.84YY535 pKa = 8.5PVEE538 pKa = 5.12LGPMDD543 pKa = 4.43NGDD546 pKa = 3.92SEE548 pKa = 5.18EE549 pKa = 4.54GLLQVHH555 pKa = 6.86DD556 pKa = 5.72AITVLNGPDD565 pKa = 4.62DD566 pKa = 6.03DD567 pKa = 4.11IASCFTVFSRR577 pKa = 11.84PSWAKK582 pKa = 8.98QADD585 pKa = 3.9RR586 pKa = 11.84KK587 pKa = 10.3RR588 pKa = 11.84RR589 pKa = 11.84EE590 pKa = 4.1AQEE593 pKa = 3.91VMAA596 pKa = 6.07
MM1 pKa = 7.72PKK3 pKa = 10.27DD4 pKa = 3.49ISLDD8 pKa = 4.05DD9 pKa = 3.59IKK11 pKa = 11.1EE12 pKa = 3.95IALRR16 pKa = 11.84AYY18 pKa = 10.17RR19 pKa = 11.84GGEE22 pKa = 3.84VRR24 pKa = 11.84SAYY27 pKa = 9.99VDD29 pKa = 3.46KK30 pKa = 11.45SDD32 pKa = 4.22DD33 pKa = 3.4NSLISEE39 pKa = 4.84EE40 pKa = 4.18KK41 pKa = 10.06TEE43 pKa = 4.38LPPEE47 pKa = 4.41SVDD50 pKa = 3.68NPEE53 pKa = 3.86EE54 pKa = 3.91NAPEE58 pKa = 4.03SEE60 pKa = 4.33EE61 pKa = 3.98TVEE64 pKa = 4.08AVEE67 pKa = 5.09DD68 pKa = 3.8ATTVTDD74 pKa = 5.63SPDD77 pKa = 3.76CDD79 pKa = 3.86TQSSEE84 pKa = 4.6SEE86 pKa = 3.94EE87 pKa = 4.12TKK89 pKa = 11.05ARR91 pKa = 11.84DD92 pKa = 3.54WPDD95 pKa = 3.27LEE97 pKa = 5.78LDD99 pKa = 3.12IARR102 pKa = 11.84ALWEE106 pKa = 4.98GEE108 pKa = 4.23LHH110 pKa = 6.23WEE112 pKa = 5.11FIPSEE117 pKa = 4.15VEE119 pKa = 3.26QWARR123 pKa = 11.84KK124 pKa = 6.06MVKK127 pKa = 9.99RR128 pKa = 11.84RR129 pKa = 11.84DD130 pKa = 3.53GEE132 pKa = 4.07MMRR135 pKa = 11.84WLLVLQQVPDD145 pKa = 3.17ILSRR149 pKa = 11.84SRR151 pKa = 11.84EE152 pKa = 3.99FIFKK156 pKa = 10.1VIRR159 pKa = 11.84SPHH162 pKa = 6.26DD163 pKa = 3.26PEE165 pKa = 6.47IYY167 pKa = 8.87LTPEE171 pKa = 4.66AIDD174 pKa = 4.22AYY176 pKa = 9.96VASFVEE182 pKa = 4.2QEE184 pKa = 4.41TVEE187 pKa = 4.07EE188 pKa = 4.61VPWSEE193 pKa = 4.43EE194 pKa = 3.49ATADD198 pKa = 3.28IAEE201 pKa = 4.37TLTLEE206 pKa = 4.8RR207 pKa = 11.84DD208 pKa = 3.57AQRR211 pKa = 11.84LAKK214 pKa = 10.04FLEE217 pKa = 4.68PEE219 pKa = 4.17AVDD222 pKa = 4.66LPEE225 pKa = 6.35GPDD228 pKa = 4.9ADD230 pKa = 4.86DD231 pKa = 3.36EE232 pKa = 4.55WVPGVPEE239 pKa = 4.68GEE241 pKa = 4.34PEE243 pKa = 4.14TTVEE247 pKa = 4.91SEE249 pKa = 4.07TADD252 pKa = 3.47APDD255 pKa = 3.17IAEE258 pKa = 4.67PEE260 pKa = 4.42TVDD263 pKa = 4.52AEE265 pKa = 4.28ATEE268 pKa = 4.25SEE270 pKa = 4.65IYY272 pKa = 10.52YY273 pKa = 10.55SPEE276 pKa = 3.25TDD278 pKa = 2.95MPTFLRR284 pKa = 11.84DD285 pKa = 3.41LTPVNGAEE293 pKa = 3.81EE294 pKa = 4.45DD295 pKa = 3.59GRR297 pKa = 11.84LFATVDD303 pKa = 3.39ALNEE307 pKa = 3.97RR308 pKa = 11.84LAGIPAGGSLVLWGLDD324 pKa = 2.87NGLNHH329 pKa = 6.57SADD332 pKa = 4.5GYY334 pKa = 10.77SSTQIRR340 pKa = 11.84IALRR344 pKa = 11.84RR345 pKa = 11.84GKK347 pKa = 9.32PAATYY352 pKa = 8.87WYY354 pKa = 9.2KK355 pKa = 10.71DD356 pKa = 3.12APRR359 pKa = 11.84RR360 pKa = 11.84QEE362 pKa = 3.58DD363 pKa = 3.3SRR365 pKa = 11.84ALFIGSAVHH374 pKa = 6.81AALLEE379 pKa = 4.1PEE381 pKa = 4.47RR382 pKa = 11.84FATEE386 pKa = 4.02YY387 pKa = 11.31ACAPSVNLRR396 pKa = 11.84TSEE399 pKa = 4.01GKK401 pKa = 8.05EE402 pKa = 3.66TLASFEE408 pKa = 4.51CDD410 pKa = 2.88CATRR414 pKa = 11.84NLIPIKK420 pKa = 10.58RR421 pKa = 11.84EE422 pKa = 3.97DD423 pKa = 3.9FDD425 pKa = 4.7TVCMMRR431 pKa = 11.84DD432 pKa = 3.26SVLAFPVVRR441 pKa = 11.84LLLSSGVGEE450 pKa = 4.72LSVFWRR456 pKa = 11.84MEE458 pKa = 3.64NGLLLKK464 pKa = 10.12IRR466 pKa = 11.84PDD468 pKa = 3.1WLGVYY473 pKa = 9.91QDD475 pKa = 4.44APFMLDD481 pKa = 3.52VKK483 pKa = 10.28TLDD486 pKa = 3.75DD487 pKa = 3.88VNDD490 pKa = 5.01FGRR493 pKa = 11.84QAHH496 pKa = 4.86EE497 pKa = 4.08HH498 pKa = 6.64GYY500 pKa = 10.09HH501 pKa = 5.8IQAAYY506 pKa = 9.87YY507 pKa = 10.23GFVLEE512 pKa = 5.15HH513 pKa = 5.99VFGMVINGLFCTIGKK528 pKa = 8.5RR529 pKa = 11.84TEE531 pKa = 3.86CGRR534 pKa = 11.84YY535 pKa = 8.5PVEE538 pKa = 5.12LGPMDD543 pKa = 4.43NGDD546 pKa = 3.92SEE548 pKa = 5.18EE549 pKa = 4.54GLLQVHH555 pKa = 6.86DD556 pKa = 5.72AITVLNGPDD565 pKa = 4.62DD566 pKa = 6.03DD567 pKa = 4.11IASCFTVFSRR577 pKa = 11.84PSWAKK582 pKa = 8.98QADD585 pKa = 3.9RR586 pKa = 11.84KK587 pKa = 10.3RR588 pKa = 11.84RR589 pKa = 11.84EE590 pKa = 4.1AQEE593 pKa = 3.91VMAA596 pKa = 6.07
Molecular weight: 66.75 kDa
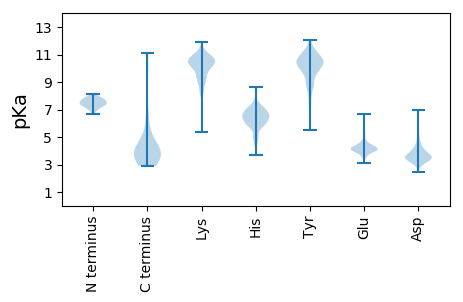
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A3E2L4|A3E2L4_9CAUD Gp55 OS=Sodalis phage phiSG1 OX=373126 PE=3 SV=1
MM1 pKa = 7.53AMKK4 pKa = 10.3DD5 pKa = 2.88KK6 pKa = 10.99SYY8 pKa = 10.37AQRR11 pKa = 11.84RR12 pKa = 11.84FEE14 pKa = 3.76RR15 pKa = 11.84TRR17 pKa = 11.84AHH19 pKa = 6.63FEE21 pKa = 4.09KK22 pKa = 10.73INDD25 pKa = 3.52DD26 pKa = 3.34RR27 pKa = 11.84ALARR31 pKa = 11.84KK32 pKa = 9.08VSKK35 pKa = 10.26AWAKK39 pKa = 8.53ITQPEE44 pKa = 4.16QTLKK48 pKa = 10.78AYY50 pKa = 9.43RR51 pKa = 11.84QQIIDD56 pKa = 3.18ATEE59 pKa = 3.96RR60 pKa = 11.84YY61 pKa = 8.6NALKK65 pKa = 10.54KK66 pKa = 9.12SARR69 pKa = 11.84YY70 pKa = 9.69RR71 pKa = 11.84MAIARR76 pKa = 4.05
MM1 pKa = 7.53AMKK4 pKa = 10.3DD5 pKa = 2.88KK6 pKa = 10.99SYY8 pKa = 10.37AQRR11 pKa = 11.84RR12 pKa = 11.84FEE14 pKa = 3.76RR15 pKa = 11.84TRR17 pKa = 11.84AHH19 pKa = 6.63FEE21 pKa = 4.09KK22 pKa = 10.73INDD25 pKa = 3.52DD26 pKa = 3.34RR27 pKa = 11.84ALARR31 pKa = 11.84KK32 pKa = 9.08VSKK35 pKa = 10.26AWAKK39 pKa = 8.53ITQPEE44 pKa = 4.16QTLKK48 pKa = 10.78AYY50 pKa = 9.43RR51 pKa = 11.84QQIIDD56 pKa = 3.18ATEE59 pKa = 3.96RR60 pKa = 11.84YY61 pKa = 8.6NALKK65 pKa = 10.54KK66 pKa = 9.12SARR69 pKa = 11.84YY70 pKa = 9.69RR71 pKa = 11.84MAIARR76 pKa = 4.05
Molecular weight: 9.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12786 |
71 |
724 |
250.7 |
27.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.48 ± 0.688 | 1.22 ± 0.151 |
5.85 ± 0.328 | 6.796 ± 0.364 |
3.582 ± 0.22 | 7.164 ± 0.34 |
2.112 ± 0.21 | 5.279 ± 0.259 |
5.529 ± 0.331 | 8.376 ± 0.285 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.464 ± 0.148 | 4.012 ± 0.283 |
4.106 ± 0.345 | 4.974 ± 0.409 |
6.499 ± 0.405 | 5.608 ± 0.254 |
5.545 ± 0.212 | 6.233 ± 0.271 |
1.251 ± 0.122 | 2.917 ± 0.234 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
