
Lishi Spider Virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Lispiviridae; Arlivirus; Lishi arlivirus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
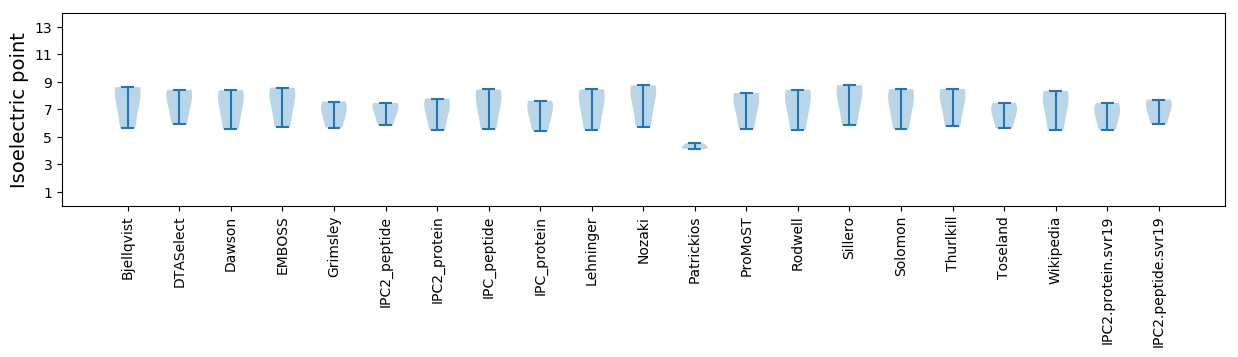
Summary statistics related to proteome-wise predictions

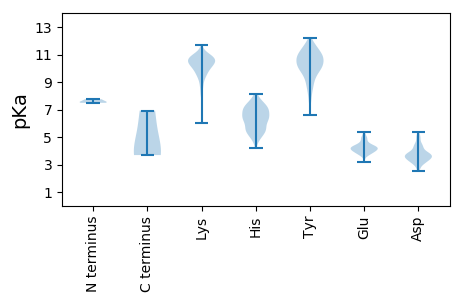
Protein with the lowest isoelectric point:
>tr|A0A0B5KRF3|A0A0B5KRF3_9MONO RNA-directed RNA polymerase L OS=Lishi Spider Virus 2 OX=1608058 GN=L PE=3 SV=1
MM1 pKa = 7.75CSNFKK6 pKa = 10.83KK7 pKa = 10.74FGICSNGMRR16 pKa = 11.84CKK18 pKa = 10.4KK19 pKa = 10.09FHH21 pKa = 7.56DD22 pKa = 4.19YY23 pKa = 11.56EE24 pKa = 4.43EE25 pKa = 5.38DD26 pKa = 3.53NLDD29 pKa = 3.43IQVGLNLIMGAISGISEE46 pKa = 4.17KK47 pKa = 10.56IKK49 pKa = 10.87NIEE52 pKa = 3.88NKK54 pKa = 9.47INEE57 pKa = 4.34IEE59 pKa = 4.27VVMSQKK65 pKa = 10.6PKK67 pKa = 10.37FQEE70 pKa = 4.13FPNNSDD76 pKa = 3.49SLNSRR81 pKa = 11.84LNMISEE87 pKa = 4.14QLDD90 pKa = 4.0TIKK93 pKa = 11.0GSIHH97 pKa = 5.13TQGIPLSTTFSNFRR111 pKa = 11.84GKK113 pKa = 9.56WSTTQNTNPKK123 pKa = 9.47GNKK126 pKa = 8.14TKK128 pKa = 10.8SSLNEE133 pKa = 3.71RR134 pKa = 11.84SIDD137 pKa = 3.67LYY139 pKa = 11.32EE140 pKa = 4.88KK141 pKa = 10.5IPAKK145 pKa = 10.03GAKK148 pKa = 10.06SMMNKK153 pKa = 9.52KK154 pKa = 10.32VSGKK158 pKa = 8.09EE159 pKa = 3.74AEE161 pKa = 4.15NRR163 pKa = 11.84TIRR166 pKa = 11.84NDD168 pKa = 3.1SHH170 pKa = 7.27EE171 pKa = 4.52SDD173 pKa = 4.51SDD175 pKa = 3.77NTSKK179 pKa = 11.16NSGGSFIKK187 pKa = 10.48EE188 pKa = 3.84LVSVQIEE195 pKa = 4.55SQSGTSEE202 pKa = 4.05SPEE205 pKa = 3.67LSGEE209 pKa = 4.16RR210 pKa = 11.84FQKK213 pKa = 10.68PFMNMEE219 pKa = 4.88LAPPLSPTGSCEE231 pKa = 4.15HH232 pKa = 6.83MEE234 pKa = 4.3TEE236 pKa = 5.18NPPEE240 pKa = 4.19HH241 pKa = 7.25QGMTDD246 pKa = 3.05NDD248 pKa = 3.7YY249 pKa = 11.63SDD251 pKa = 5.02DD252 pKa = 3.53NRR254 pKa = 11.84KK255 pKa = 9.23IFTQKK260 pKa = 9.23TPAIEE265 pKa = 4.14SFKK268 pKa = 10.99KK269 pKa = 10.37DD270 pKa = 3.5DD271 pKa = 4.29EE272 pKa = 4.93LDD274 pKa = 3.09IHH276 pKa = 6.92
MM1 pKa = 7.75CSNFKK6 pKa = 10.83KK7 pKa = 10.74FGICSNGMRR16 pKa = 11.84CKK18 pKa = 10.4KK19 pKa = 10.09FHH21 pKa = 7.56DD22 pKa = 4.19YY23 pKa = 11.56EE24 pKa = 4.43EE25 pKa = 5.38DD26 pKa = 3.53NLDD29 pKa = 3.43IQVGLNLIMGAISGISEE46 pKa = 4.17KK47 pKa = 10.56IKK49 pKa = 10.87NIEE52 pKa = 3.88NKK54 pKa = 9.47INEE57 pKa = 4.34IEE59 pKa = 4.27VVMSQKK65 pKa = 10.6PKK67 pKa = 10.37FQEE70 pKa = 4.13FPNNSDD76 pKa = 3.49SLNSRR81 pKa = 11.84LNMISEE87 pKa = 4.14QLDD90 pKa = 4.0TIKK93 pKa = 11.0GSIHH97 pKa = 5.13TQGIPLSTTFSNFRR111 pKa = 11.84GKK113 pKa = 9.56WSTTQNTNPKK123 pKa = 9.47GNKK126 pKa = 8.14TKK128 pKa = 10.8SSLNEE133 pKa = 3.71RR134 pKa = 11.84SIDD137 pKa = 3.67LYY139 pKa = 11.32EE140 pKa = 4.88KK141 pKa = 10.5IPAKK145 pKa = 10.03GAKK148 pKa = 10.06SMMNKK153 pKa = 9.52KK154 pKa = 10.32VSGKK158 pKa = 8.09EE159 pKa = 3.74AEE161 pKa = 4.15NRR163 pKa = 11.84TIRR166 pKa = 11.84NDD168 pKa = 3.1SHH170 pKa = 7.27EE171 pKa = 4.52SDD173 pKa = 4.51SDD175 pKa = 3.77NTSKK179 pKa = 11.16NSGGSFIKK187 pKa = 10.48EE188 pKa = 3.84LVSVQIEE195 pKa = 4.55SQSGTSEE202 pKa = 4.05SPEE205 pKa = 3.67LSGEE209 pKa = 4.16RR210 pKa = 11.84FQKK213 pKa = 10.68PFMNMEE219 pKa = 4.88LAPPLSPTGSCEE231 pKa = 4.15HH232 pKa = 6.83MEE234 pKa = 4.3TEE236 pKa = 5.18NPPEE240 pKa = 4.19HH241 pKa = 7.25QGMTDD246 pKa = 3.05NDD248 pKa = 3.7YY249 pKa = 11.63SDD251 pKa = 5.02DD252 pKa = 3.53NRR254 pKa = 11.84KK255 pKa = 9.23IFTQKK260 pKa = 9.23TPAIEE265 pKa = 4.14SFKK268 pKa = 10.99KK269 pKa = 10.37DD270 pKa = 3.5DD271 pKa = 4.29EE272 pKa = 4.93LDD274 pKa = 3.09IHH276 pKa = 6.92
Molecular weight: 30.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KK20|A0A0B5KK20_9MONO ORF2 OS=Lishi Spider Virus 2 OX=1608058 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.55SKK3 pKa = 10.2FRR5 pKa = 11.84NILIIFISMLSPGLFIEE22 pKa = 5.32RR23 pKa = 11.84YY24 pKa = 9.33HH25 pKa = 6.87PIFFEE30 pKa = 4.3KK31 pKa = 10.64YY32 pKa = 9.21GVLIEE37 pKa = 4.36PNGVVTSYY45 pKa = 11.8DD46 pKa = 2.99KK47 pKa = 11.35VKK49 pKa = 10.24YY50 pKa = 8.89VTLIYY55 pKa = 10.24HH56 pKa = 6.47FPGLQKK62 pKa = 10.76AVTPSHH68 pKa = 6.75CNRR71 pKa = 11.84NASEE75 pKa = 3.74QLYY78 pKa = 10.68RR79 pKa = 11.84NTLKK83 pKa = 10.75HH84 pKa = 6.01QIKK87 pKa = 10.16IMYY90 pKa = 9.49DD91 pKa = 3.79LIPSSTHH98 pKa = 6.0TIMSNYY104 pKa = 8.9CSKK107 pKa = 10.62HH108 pKa = 5.55HH109 pKa = 6.55ALCLDD114 pKa = 4.03LEE116 pKa = 4.69KK117 pKa = 10.52KK118 pKa = 10.11DD119 pKa = 3.59RR120 pKa = 11.84PKK122 pKa = 10.75RR123 pKa = 11.84QIIAAIAALSGIAGLAISAYY143 pKa = 10.02DD144 pKa = 3.65WISSSEE150 pKa = 4.14LKK152 pKa = 10.67NHH154 pKa = 6.39LDD156 pKa = 3.35KK157 pKa = 11.47VSEE160 pKa = 4.9HH161 pKa = 5.47ITKK164 pKa = 9.74IDD166 pKa = 3.32NKK168 pKa = 10.93LIIQEE173 pKa = 4.02NLDD176 pKa = 3.63YY177 pKa = 11.34KK178 pKa = 9.96LTKK181 pKa = 10.34LSNDD185 pKa = 3.63IIKK188 pKa = 8.48KK189 pKa = 8.37TSMSFEE195 pKa = 4.3NYY197 pKa = 8.99HH198 pKa = 6.35KK199 pKa = 10.32FVKK202 pKa = 10.37EE203 pKa = 3.83YY204 pKa = 10.61VCQSSQFNNFLSHH217 pKa = 6.83HH218 pKa = 6.07VATTHH223 pKa = 7.71LEE225 pKa = 4.06MEE227 pKa = 4.39LQALQRR233 pKa = 11.84LINGEE238 pKa = 3.93PDD240 pKa = 3.21SYY242 pKa = 11.01IINAEE247 pKa = 4.05TLEE250 pKa = 4.9SILSSDD256 pKa = 3.86TEE258 pKa = 4.4VEE260 pKa = 3.88RR261 pKa = 11.84SIYY264 pKa = 10.85SSDD267 pKa = 3.21PSLFYY272 pKa = 10.6QVVRR276 pKa = 11.84SNLIHH281 pKa = 7.11SDD283 pKa = 3.1IEE285 pKa = 3.95KK286 pKa = 10.85SLFAFILEE294 pKa = 4.12VPIIQEE300 pKa = 4.14TMISPLYY307 pKa = 10.08KK308 pKa = 10.05VYY310 pKa = 11.12NGGWEE315 pKa = 4.3SKK317 pKa = 10.89GILHH321 pKa = 7.37KK322 pKa = 10.23IDD324 pKa = 4.83LPDD327 pKa = 3.2SFYY330 pKa = 10.97LYY332 pKa = 10.96SSLDD336 pKa = 3.52DD337 pKa = 3.56VDD339 pKa = 3.71YY340 pKa = 11.13HH341 pKa = 7.08AISLGKK347 pKa = 9.53SNCWKK352 pKa = 9.59RR353 pKa = 11.84NEE355 pKa = 4.49VVVCDD360 pKa = 4.0NSKK363 pKa = 10.92HH364 pKa = 5.36MMTEE368 pKa = 4.82DD369 pKa = 4.4MICLNSILKK378 pKa = 10.12RR379 pKa = 11.84EE380 pKa = 4.32YY381 pKa = 10.94YY382 pKa = 10.31DD383 pKa = 3.27SCDD386 pKa = 2.99IKK388 pKa = 10.75FSKK391 pKa = 9.08FTRR394 pKa = 11.84RR395 pKa = 11.84KK396 pKa = 8.94FVVKK400 pKa = 10.19ALSGVLVCGDD410 pKa = 4.51LEE412 pKa = 4.57VKK414 pKa = 10.32IITSAGSQFHH424 pKa = 6.23YY425 pKa = 10.45IKK427 pKa = 10.89NNKK430 pKa = 8.03QEE432 pKa = 4.07NNYY435 pKa = 8.38TKK437 pKa = 10.24YY438 pKa = 10.79YY439 pKa = 9.9SYY441 pKa = 11.86NEE443 pKa = 3.89FKK445 pKa = 10.87QIIVGNTIVSTNINKK460 pKa = 8.56MPIVRR465 pKa = 11.84RR466 pKa = 11.84NDD468 pKa = 3.81TISVNFDD475 pKa = 3.49YY476 pKa = 10.96DD477 pKa = 4.29LKK479 pKa = 10.16WADD482 pKa = 4.0KK483 pKa = 10.29ILHH486 pKa = 5.44EE487 pKa = 5.16RR488 pKa = 11.84PWTSLKK494 pKa = 10.43EE495 pKa = 3.91INLLKK500 pKa = 10.86DD501 pKa = 3.2SGFEE505 pKa = 4.04QYY507 pKa = 10.7HH508 pKa = 5.05GTISHH513 pKa = 6.97NLNQWSVFLWVTIILCLIILIYY535 pKa = 10.71LLIASRR541 pKa = 11.84RR542 pKa = 11.84KK543 pKa = 8.39ITSLEE548 pKa = 4.0DD549 pKa = 3.22RR550 pKa = 11.84LKK552 pKa = 10.13LTNGIIKK559 pKa = 10.49LSNKK563 pKa = 9.79RR564 pKa = 11.84SILL567 pKa = 3.69
MM1 pKa = 7.55SKK3 pKa = 10.2FRR5 pKa = 11.84NILIIFISMLSPGLFIEE22 pKa = 5.32RR23 pKa = 11.84YY24 pKa = 9.33HH25 pKa = 6.87PIFFEE30 pKa = 4.3KK31 pKa = 10.64YY32 pKa = 9.21GVLIEE37 pKa = 4.36PNGVVTSYY45 pKa = 11.8DD46 pKa = 2.99KK47 pKa = 11.35VKK49 pKa = 10.24YY50 pKa = 8.89VTLIYY55 pKa = 10.24HH56 pKa = 6.47FPGLQKK62 pKa = 10.76AVTPSHH68 pKa = 6.75CNRR71 pKa = 11.84NASEE75 pKa = 3.74QLYY78 pKa = 10.68RR79 pKa = 11.84NTLKK83 pKa = 10.75HH84 pKa = 6.01QIKK87 pKa = 10.16IMYY90 pKa = 9.49DD91 pKa = 3.79LIPSSTHH98 pKa = 6.0TIMSNYY104 pKa = 8.9CSKK107 pKa = 10.62HH108 pKa = 5.55HH109 pKa = 6.55ALCLDD114 pKa = 4.03LEE116 pKa = 4.69KK117 pKa = 10.52KK118 pKa = 10.11DD119 pKa = 3.59RR120 pKa = 11.84PKK122 pKa = 10.75RR123 pKa = 11.84QIIAAIAALSGIAGLAISAYY143 pKa = 10.02DD144 pKa = 3.65WISSSEE150 pKa = 4.14LKK152 pKa = 10.67NHH154 pKa = 6.39LDD156 pKa = 3.35KK157 pKa = 11.47VSEE160 pKa = 4.9HH161 pKa = 5.47ITKK164 pKa = 9.74IDD166 pKa = 3.32NKK168 pKa = 10.93LIIQEE173 pKa = 4.02NLDD176 pKa = 3.63YY177 pKa = 11.34KK178 pKa = 9.96LTKK181 pKa = 10.34LSNDD185 pKa = 3.63IIKK188 pKa = 8.48KK189 pKa = 8.37TSMSFEE195 pKa = 4.3NYY197 pKa = 8.99HH198 pKa = 6.35KK199 pKa = 10.32FVKK202 pKa = 10.37EE203 pKa = 3.83YY204 pKa = 10.61VCQSSQFNNFLSHH217 pKa = 6.83HH218 pKa = 6.07VATTHH223 pKa = 7.71LEE225 pKa = 4.06MEE227 pKa = 4.39LQALQRR233 pKa = 11.84LINGEE238 pKa = 3.93PDD240 pKa = 3.21SYY242 pKa = 11.01IINAEE247 pKa = 4.05TLEE250 pKa = 4.9SILSSDD256 pKa = 3.86TEE258 pKa = 4.4VEE260 pKa = 3.88RR261 pKa = 11.84SIYY264 pKa = 10.85SSDD267 pKa = 3.21PSLFYY272 pKa = 10.6QVVRR276 pKa = 11.84SNLIHH281 pKa = 7.11SDD283 pKa = 3.1IEE285 pKa = 3.95KK286 pKa = 10.85SLFAFILEE294 pKa = 4.12VPIIQEE300 pKa = 4.14TMISPLYY307 pKa = 10.08KK308 pKa = 10.05VYY310 pKa = 11.12NGGWEE315 pKa = 4.3SKK317 pKa = 10.89GILHH321 pKa = 7.37KK322 pKa = 10.23IDD324 pKa = 4.83LPDD327 pKa = 3.2SFYY330 pKa = 10.97LYY332 pKa = 10.96SSLDD336 pKa = 3.52DD337 pKa = 3.56VDD339 pKa = 3.71YY340 pKa = 11.13HH341 pKa = 7.08AISLGKK347 pKa = 9.53SNCWKK352 pKa = 9.59RR353 pKa = 11.84NEE355 pKa = 4.49VVVCDD360 pKa = 4.0NSKK363 pKa = 10.92HH364 pKa = 5.36MMTEE368 pKa = 4.82DD369 pKa = 4.4MICLNSILKK378 pKa = 10.12RR379 pKa = 11.84EE380 pKa = 4.32YY381 pKa = 10.94YY382 pKa = 10.31DD383 pKa = 3.27SCDD386 pKa = 2.99IKK388 pKa = 10.75FSKK391 pKa = 9.08FTRR394 pKa = 11.84RR395 pKa = 11.84KK396 pKa = 8.94FVVKK400 pKa = 10.19ALSGVLVCGDD410 pKa = 4.51LEE412 pKa = 4.57VKK414 pKa = 10.32IITSAGSQFHH424 pKa = 6.23YY425 pKa = 10.45IKK427 pKa = 10.89NNKK430 pKa = 8.03QEE432 pKa = 4.07NNYY435 pKa = 8.38TKK437 pKa = 10.24YY438 pKa = 10.79YY439 pKa = 9.9SYY441 pKa = 11.86NEE443 pKa = 3.89FKK445 pKa = 10.87QIIVGNTIVSTNINKK460 pKa = 8.56MPIVRR465 pKa = 11.84RR466 pKa = 11.84NDD468 pKa = 3.81TISVNFDD475 pKa = 3.49YY476 pKa = 10.96DD477 pKa = 4.29LKK479 pKa = 10.16WADD482 pKa = 4.0KK483 pKa = 10.29ILHH486 pKa = 5.44EE487 pKa = 5.16RR488 pKa = 11.84PWTSLKK494 pKa = 10.43EE495 pKa = 3.91INLLKK500 pKa = 10.86DD501 pKa = 3.2SGFEE505 pKa = 4.04QYY507 pKa = 10.7HH508 pKa = 5.05GTISHH513 pKa = 6.97NLNQWSVFLWVTIILCLIILIYY535 pKa = 10.71LLIASRR541 pKa = 11.84RR542 pKa = 11.84KK543 pKa = 8.39ITSLEE548 pKa = 4.0DD549 pKa = 3.22RR550 pKa = 11.84LKK552 pKa = 10.13LTNGIIKK559 pKa = 10.49LSNKK563 pKa = 9.79RR564 pKa = 11.84SILL567 pKa = 3.69
Molecular weight: 65.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3044 |
276 |
2201 |
1014.7 |
117.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.365 ± 0.319 | 1.708 ± 0.062 |
5.519 ± 0.128 | 6.603 ± 0.599 |
4.698 ± 0.322 | 4.008 ± 0.514 |
2.497 ± 0.385 | 9.625 ± 0.568 |
8.279 ± 0.386 | 10.414 ± 1.366 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.201 ± 0.43 | 7.096 ± 0.468 |
3.187 ± 0.373 | 3.318 ± 0.189 |
4.172 ± 0.456 | 9.428 ± 0.878 |
5.158 ± 0.201 | 4.402 ± 0.534 |
1.084 ± 0.172 | 4.238 ± 0.747 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
