
Desulfosporosinus meridiei (strain ATCC BAA-275 / DSM 13257 / KCTC 12902 / NCIMB 13706 / S10)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Peptococcaceae; Desulfosporosinus; Desulfosporosinus meridiei
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
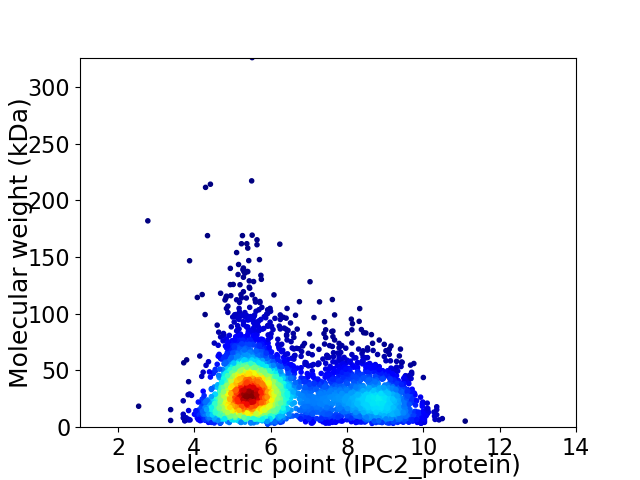
Virtual 2D-PAGE plot for 4340 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
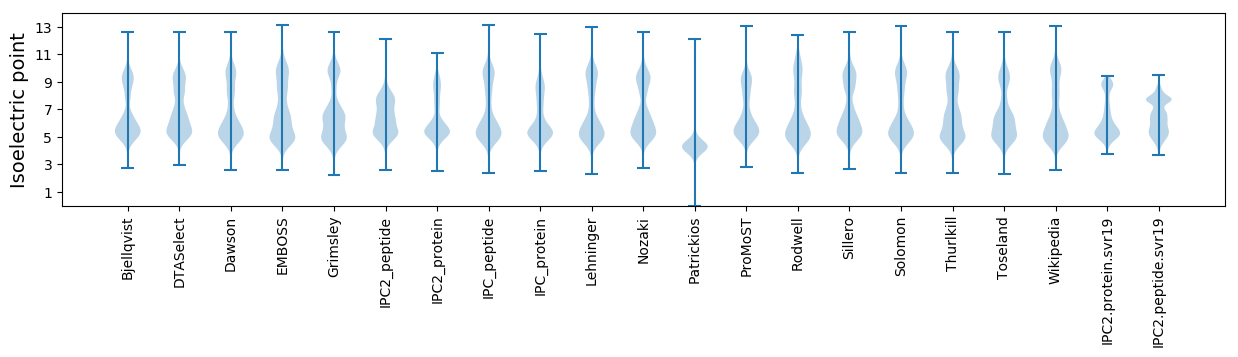
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J7IME2|J7IME2_DESMD Cation diffusion facilitator family transporter OS=Desulfosporosinus meridiei (strain ATCC BAA-275 / DSM 13257 / KCTC 12902 / NCIMB 13706 / S10) OX=768704 GN=Desmer_0943 PE=4 SV=1
MM1 pKa = 7.59FKK3 pKa = 10.03TKK5 pKa = 10.44QKK7 pKa = 10.63ILGLVMSMALGLTFFVAPAPAAAATDD33 pKa = 3.96MLTLANSDD41 pKa = 3.53HH42 pKa = 5.85FTILGEE48 pKa = 4.12VDD50 pKa = 5.06DD51 pKa = 4.94EE52 pKa = 4.72VSNIQVVGLDD62 pKa = 3.19GTTWMTEE69 pKa = 3.94EE70 pKa = 4.25IADD73 pKa = 4.05PEE75 pKa = 4.4NIEE78 pKa = 4.04WTTSDD83 pKa = 3.17SSVVKK88 pKa = 10.38FLDD91 pKa = 3.62GTTEE95 pKa = 3.77VATIDD100 pKa = 3.55DD101 pKa = 4.1TDD103 pKa = 3.79TVKK106 pKa = 10.64IKK108 pKa = 10.94LLDD111 pKa = 3.49EE112 pKa = 4.03GRR114 pKa = 11.84AYY116 pKa = 9.03VTAHH120 pKa = 6.3YY121 pKa = 11.03DD122 pKa = 3.21SMEE125 pKa = 3.5ISAYY129 pKa = 10.46VVVEE133 pKa = 4.1TDD135 pKa = 3.62GSATPSISGISVVVDD150 pKa = 3.76APGTANDD157 pKa = 4.0FTATNQTVYY166 pKa = 10.76LADD169 pKa = 4.6LSWLTDD175 pKa = 3.88DD176 pKa = 5.86SKK178 pKa = 10.14TLQKK182 pKa = 10.86NSSALHH188 pKa = 5.65ALAMAGNTNYY198 pKa = 10.39SDD200 pKa = 4.65PDD202 pKa = 3.33WAEE205 pKa = 3.7NNLTVFSGGSYY216 pKa = 10.52VYY218 pKa = 10.99GIGSDD223 pKa = 3.87FASGTDD229 pKa = 2.72GWQYY233 pKa = 11.32HH234 pKa = 4.94VVHH237 pKa = 6.71SNSTTDD243 pKa = 3.31VPAYY247 pKa = 9.66AASVYY252 pKa = 10.28EE253 pKa = 4.33LTSGDD258 pKa = 3.83TVVWEE263 pKa = 4.13YY264 pKa = 11.44KK265 pKa = 10.29GWW267 pKa = 3.56
MM1 pKa = 7.59FKK3 pKa = 10.03TKK5 pKa = 10.44QKK7 pKa = 10.63ILGLVMSMALGLTFFVAPAPAAAATDD33 pKa = 3.96MLTLANSDD41 pKa = 3.53HH42 pKa = 5.85FTILGEE48 pKa = 4.12VDD50 pKa = 5.06DD51 pKa = 4.94EE52 pKa = 4.72VSNIQVVGLDD62 pKa = 3.19GTTWMTEE69 pKa = 3.94EE70 pKa = 4.25IADD73 pKa = 4.05PEE75 pKa = 4.4NIEE78 pKa = 4.04WTTSDD83 pKa = 3.17SSVVKK88 pKa = 10.38FLDD91 pKa = 3.62GTTEE95 pKa = 3.77VATIDD100 pKa = 3.55DD101 pKa = 4.1TDD103 pKa = 3.79TVKK106 pKa = 10.64IKK108 pKa = 10.94LLDD111 pKa = 3.49EE112 pKa = 4.03GRR114 pKa = 11.84AYY116 pKa = 9.03VTAHH120 pKa = 6.3YY121 pKa = 11.03DD122 pKa = 3.21SMEE125 pKa = 3.5ISAYY129 pKa = 10.46VVVEE133 pKa = 4.1TDD135 pKa = 3.62GSATPSISGISVVVDD150 pKa = 3.76APGTANDD157 pKa = 4.0FTATNQTVYY166 pKa = 10.76LADD169 pKa = 4.6LSWLTDD175 pKa = 3.88DD176 pKa = 5.86SKK178 pKa = 10.14TLQKK182 pKa = 10.86NSSALHH188 pKa = 5.65ALAMAGNTNYY198 pKa = 10.39SDD200 pKa = 4.65PDD202 pKa = 3.33WAEE205 pKa = 3.7NNLTVFSGGSYY216 pKa = 10.52VYY218 pKa = 10.99GIGSDD223 pKa = 3.87FASGTDD229 pKa = 2.72GWQYY233 pKa = 11.32HH234 pKa = 4.94VVHH237 pKa = 6.71SNSTTDD243 pKa = 3.31VPAYY247 pKa = 9.66AASVYY252 pKa = 10.28EE253 pKa = 4.33LTSGDD258 pKa = 3.83TVVWEE263 pKa = 4.13YY264 pKa = 11.44KK265 pKa = 10.29GWW267 pKa = 3.56
Molecular weight: 28.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J7ITW7|J7ITW7_DESMD ABC-type dipeptide/oligopeptide/nickel transport system ATPase component OS=Desulfosporosinus meridiei (strain ATCC BAA-275 / DSM 13257 / KCTC 12902 / NCIMB 13706 / S10) OX=768704 GN=Desmer_0519 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 9.19QPKK8 pKa = 8.78NRR10 pKa = 11.84RR11 pKa = 11.84HH12 pKa = 5.44KK13 pKa = 10.09RR14 pKa = 11.84VHH16 pKa = 5.93GFLSRR21 pKa = 11.84MSTPTGRR28 pKa = 11.84NVIKK32 pKa = 10.5RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.52GRR39 pKa = 11.84KK40 pKa = 8.8KK41 pKa = 10.81LSVV44 pKa = 3.15
MM1 pKa = 7.36KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 9.19QPKK8 pKa = 8.78NRR10 pKa = 11.84RR11 pKa = 11.84HH12 pKa = 5.44KK13 pKa = 10.09RR14 pKa = 11.84VHH16 pKa = 5.93GFLSRR21 pKa = 11.84MSTPTGRR28 pKa = 11.84NVIKK32 pKa = 10.5RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.52GRR39 pKa = 11.84KK40 pKa = 8.8KK41 pKa = 10.81LSVV44 pKa = 3.15
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1328786 |
30 |
2859 |
306.2 |
34.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.562 ± 0.043 | 1.088 ± 0.016 |
4.895 ± 0.027 | 6.909 ± 0.045 |
4.096 ± 0.026 | 7.394 ± 0.056 |
1.754 ± 0.017 | 7.601 ± 0.032 |
6.219 ± 0.039 | 10.503 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.623 ± 0.019 | 4.228 ± 0.03 |
3.858 ± 0.025 | 3.76 ± 0.028 |
4.534 ± 0.03 | 6.258 ± 0.03 |
5.3 ± 0.043 | 7.112 ± 0.034 |
1.05 ± 0.014 | 3.256 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
