
Coccomyxa subellipsoidea (strain C-169) (Green microalga)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; core chlorophytes; Trebouxiophyceae; Trebouxiophyceae incertae sedis; Elliptochloris clade; Coccomyxa; Coccomyxa subellipsoidea
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
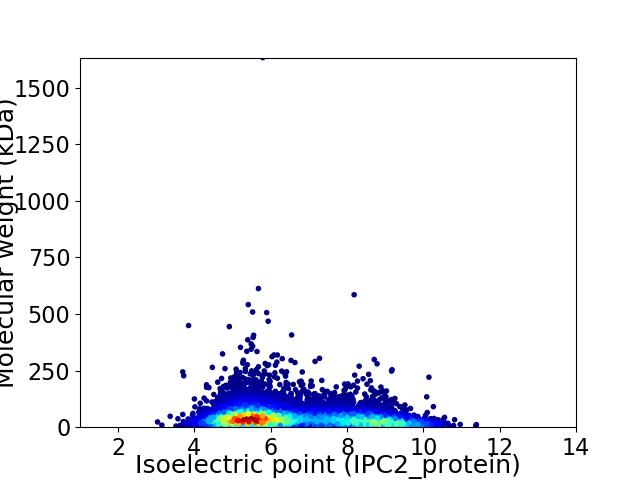
Virtual 2D-PAGE plot for 9799 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I0YSZ9|I0YSZ9_COCSC Exostosin domain-containing protein OS=Coccomyxa subellipsoidea (strain C-169) OX=574566 GN=COCSUDRAFT_66927 PE=3 SV=1
MM1 pKa = 7.68LEE3 pKa = 4.05PRR5 pKa = 11.84VLHH8 pKa = 7.17DD9 pKa = 4.05IVACTAVVQSPDD21 pKa = 3.02VLTRR25 pKa = 11.84WWLQNANTSRR35 pKa = 11.84APDD38 pKa = 3.52AGIAIPISHH47 pKa = 7.29FSSRR51 pKa = 11.84WHH53 pKa = 6.83LGDD56 pKa = 4.58GPAANDD62 pKa = 4.41GAWPAQPPSLSFAANSDD79 pKa = 3.38DD80 pKa = 3.68GGRR83 pKa = 11.84GGSGAAAPPADD94 pKa = 3.63QTLAATALASIRR106 pKa = 11.84KK107 pKa = 8.43GVTRR111 pKa = 11.84VPVTAVTPPRR121 pKa = 11.84PTSATLASTDD131 pKa = 3.2AQAAIVQQQAAPDD144 pKa = 3.98GEE146 pKa = 4.98AYY148 pKa = 10.23ASRR151 pKa = 11.84VARR154 pKa = 11.84SAARR158 pKa = 11.84VAEE161 pKa = 4.12LAGAAGDD168 pKa = 4.04STAGLMWSSALGSAFQDD185 pKa = 3.03GGAAVVNGASGAVGLADD202 pKa = 4.83DD203 pKa = 4.79AVTGAGVQAAQDD215 pKa = 3.68MSTLRR220 pKa = 11.84DD221 pKa = 3.39GLAAAVTPIANATVATAAAARR242 pKa = 11.84QALTQGIEE250 pKa = 4.01DD251 pKa = 3.91TTEE254 pKa = 3.42QFAPVLAPLRR264 pKa = 11.84PGSASEE270 pKa = 4.06ALAPAVAQIGDD281 pKa = 3.33WWRR284 pKa = 11.84QSQEE288 pKa = 3.69RR289 pKa = 11.84ARR291 pKa = 11.84SSLNDD296 pKa = 3.44FLTPALAGVANVVSPQKK313 pKa = 10.53RR314 pKa = 11.84EE315 pKa = 3.34EE316 pKa = 4.13CMQKK320 pKa = 8.79FTHH323 pKa = 6.93CGFCSPNFDD332 pKa = 3.17RR333 pKa = 11.84CLFCTAPFTLIDD345 pKa = 4.34ISCACPPGTFVSSDD359 pKa = 3.25GSSCIPFAVAQLEE372 pKa = 4.1KK373 pKa = 10.7LILAISQQNAALADD387 pKa = 3.93RR388 pKa = 11.84VNSLLHH394 pKa = 6.18LVGPSRR400 pKa = 11.84TCQANIPFCLRR411 pKa = 11.84CSSPATCAVCSDD423 pKa = 4.28GYY425 pKa = 11.47KK426 pKa = 10.23LDD428 pKa = 4.26AVGKK432 pKa = 7.93CICDD436 pKa = 3.5PTTAGPGCAFCIVPGSCGNPLGGTKK461 pKa = 10.58ANLKK465 pKa = 10.07ACADD469 pKa = 3.75FGKK472 pKa = 10.1RR473 pKa = 11.84RR474 pKa = 11.84QYY476 pKa = 9.96GRR478 pKa = 11.84AYY480 pKa = 10.81NFVKK484 pKa = 10.8DD485 pKa = 3.72PVTGLGRR492 pKa = 11.84CEE494 pKa = 4.99CNPQSTPHH502 pKa = 6.49CAACLFPGVCKK513 pKa = 10.59FCNAPFEE520 pKa = 4.52LVDD523 pKa = 4.62GKK525 pKa = 10.05CACPPLYY532 pKa = 10.25GPPVMDD538 pKa = 5.34EE539 pKa = 4.22DD540 pKa = 4.0TCLPIFLGQQVYY552 pKa = 7.79ITGPTGPTGFGPTGPTGIAIGATGFTGSTAYY583 pKa = 10.32GVTGFTGFTGFTGFTGFTGPTGFTGFTGDD612 pKa = 3.06TGFTGFTGDD621 pKa = 3.06TGFTGFTGDD630 pKa = 3.06TGFTGFTGDD639 pKa = 3.22TGFTGTTGDD648 pKa = 3.25TGFTGFTGDD657 pKa = 3.46TGFTGNTGDD666 pKa = 3.72TGFTGATGFTGDD678 pKa = 3.64TGFTGDD684 pKa = 3.77TGFTGNTGDD693 pKa = 3.72TGFTGATGFTGDD705 pKa = 3.43TGTTGFTGTTGNTGDD720 pKa = 3.7TGFTGDD726 pKa = 3.32TGFTGFTGATGDD738 pKa = 3.57TGFTGNTGDD747 pKa = 3.39TGFTGFTGNTGFTGFTGDD765 pKa = 3.31TGFTGDD771 pKa = 3.72TGFTGATGFTGDD783 pKa = 3.31TGFTGFTGSTGFTGFTGDD801 pKa = 3.06TGFTGFTGFTGATGNTGDD819 pKa = 3.62TGFTGFTGDD828 pKa = 3.06TGFTGFTGDD837 pKa = 3.06TGFTGFTGFTGDD849 pKa = 3.46TGFTGNTGDD858 pKa = 3.72TGFTGATGFSGSTGFTGDD876 pKa = 3.18TGFAGATGFTGFTGDD891 pKa = 3.42TGFTGATGVTGFTGATGYY909 pKa = 10.91SGDD912 pKa = 3.83TGFTGATGFSGFTGFTGGTGFTGFTGDD939 pKa = 3.03TGFTGFPGFTGATGFTGFTGATGFTGDD966 pKa = 3.31TGFTGFTGNTGFTGATGFTGFTGFTGDD993 pKa = 3.42TGFTGATGFTGFTGATGFTGSTGFTGDD1020 pKa = 2.93TGFTGVSGFTGDD1032 pKa = 3.32TGFTGATGFTGFTGATGFTGDD1053 pKa = 4.0TGISGATGFTGFTGFTGATGFTGTTGFTGATGLTGDD1089 pKa = 4.18SGFTGTTGFTGFTGDD1104 pKa = 3.06TGFTGFTGFTGFTGATGFTGDD1125 pKa = 4.09TGNTGATGFTGFTGATGFTGDD1146 pKa = 3.31TGFTGFTGATGFTGATGFTGDD1167 pKa = 3.31TGFTGFTGFTGFTGDD1182 pKa = 3.42TGFTGATGFTGFTGATGITGDD1203 pKa = 3.89TGITGATGFTGATGYY1218 pKa = 9.12TGFTGNTGSTGSTGATGFTGATGYY1242 pKa = 9.24TGTTGFTGFTGDD1254 pKa = 3.46TGFTGNTGDD1263 pKa = 3.43TGFTGVTGFTGATGFTGSTGDD1284 pKa = 3.36TGFTGATGFTGFTGATGFTGSTGFTGTTGFTGATGFTGFTGFTGTTGATGFTGFTGATGFTGFTGFTGATGFTGATGSTGFTGFTGDD1371 pKa = 3.5TGITGATGFTGDD1383 pKa = 3.55TGFTGTTGFTGFTGDD1398 pKa = 3.42TGFTGATGFTGTTGFTGATGFTGFTGDD1425 pKa = 3.21TGITGGTGFTGSTGFTGNTGSTGATGITGFTGATGFTGDD1464 pKa = 3.76TGFTGATGFTGFTGDD1479 pKa = 3.42TGFTGATGSTGFTGATGFTGFTGATGFTGFTGGTGYY1515 pKa = 9.2TGTTGFTGATGFTGFTGDD1533 pKa = 3.5TGITGATGFTGSTGFTGTTGFTGFTGDD1560 pKa = 3.23TGFTGSTGATGFTGATGFTGGTGFTGFTGSTGFTGAIGFTGATGFTGFTGDD1611 pKa = 3.23TGFTGSTGFTGFTGDD1626 pKa = 3.42TGFTGATGFTGATGNTGFTGATGFTGTTGFTGFTGDD1662 pKa = 3.42TGFTGATGSTGFTGSTGFSGATGFSGATGFTGFTGATGFTGDD1704 pKa = 3.39TGATGFSGATGFTGSTGGTGATGYY1728 pKa = 10.76SGFTGATGFTGDD1740 pKa = 3.82SGFTGTTGATGYY1752 pKa = 8.98TGSTGFTGATGFTGFTGATGYY1773 pKa = 9.01TGATGFTGFTGDD1785 pKa = 3.06TGFTGFTGNTGFTGATGFTGSTGATGFTGATGLTGFTGATGSTGTSGSTGFTGATGFTGVTGATGFTGSTGFTGSTGFTGSTGFTGATGSTGATGDD1881 pKa = 3.43TGFTGNTGFTGNTGFTGGTGATGYY1905 pKa = 8.99TGATGFTGFTGTTGDD1920 pKa = 3.42TGITGSTGYY1929 pKa = 8.97TGSTGATGFSGATGFTGFTGDD1950 pKa = 3.42TGFTGATGFTGSTGVTGFTGGTGYY1974 pKa = 9.11TGFTGSTGATGFTGVTGFTGDD1995 pKa = 3.06TGATGFSGATGATGVTGSTGFTGSTGFTGATGFTGDD2031 pKa = 3.7TGATGLTGASGFTGNTGFTGATGFTGFTGATGFTGFTGFTGATGFTGNTGATGASGFTGSTGGTGFTGATGFTGSTGFTGFTGDD2115 pKa = 3.64TGFSGATGFTGSTGGTGTTGATGFTGSTGGTGSSGATGATGGSGATGFTGGTGATGLTGNTGATGSTGGTGFTGTTGGTGATGDD2199 pKa = 3.63TGSTGGTGITGVTGGTGALGSTGATGFTGDD2229 pKa = 3.81TGITGATGFSGATGATGFSGPSGVTGITGATGFTGSSGDD2268 pKa = 3.53TGVTGATGSRR2278 pKa = 11.84GFIGATGFTGAIGTTGSTGFTGGTGATGGTGGTGFTGATGSTGTTGGTGGTGGTGASGSTGNTGASGFTGSTGGTGATGFTGQTGGTGSTGTTGATGSTGTTGSSGITGSTGSTGSSGLQGFTGATGATGNSGTTGATGLTGASGATGFTGSTGGSGATGSTGFSGATGTTGATGFSGGSGATGSTGFSGATGGTGSTGGTGSTGATGGTGFSGATGFTGTTGGTGFTGFSGATGGSGFTGATGAAATGATGVTGNTGPTGGTGPSGSTGAVGTSGATGSTGVTGTTGTSGPTGFSGGTGSTGPGITGATGVTGPTGPTGPTGFTGGTGGTGGTGFAATGATGPTGPTGAQGVTGGAGG2636 pKa = 2.87
MM1 pKa = 7.68LEE3 pKa = 4.05PRR5 pKa = 11.84VLHH8 pKa = 7.17DD9 pKa = 4.05IVACTAVVQSPDD21 pKa = 3.02VLTRR25 pKa = 11.84WWLQNANTSRR35 pKa = 11.84APDD38 pKa = 3.52AGIAIPISHH47 pKa = 7.29FSSRR51 pKa = 11.84WHH53 pKa = 6.83LGDD56 pKa = 4.58GPAANDD62 pKa = 4.41GAWPAQPPSLSFAANSDD79 pKa = 3.38DD80 pKa = 3.68GGRR83 pKa = 11.84GGSGAAAPPADD94 pKa = 3.63QTLAATALASIRR106 pKa = 11.84KK107 pKa = 8.43GVTRR111 pKa = 11.84VPVTAVTPPRR121 pKa = 11.84PTSATLASTDD131 pKa = 3.2AQAAIVQQQAAPDD144 pKa = 3.98GEE146 pKa = 4.98AYY148 pKa = 10.23ASRR151 pKa = 11.84VARR154 pKa = 11.84SAARR158 pKa = 11.84VAEE161 pKa = 4.12LAGAAGDD168 pKa = 4.04STAGLMWSSALGSAFQDD185 pKa = 3.03GGAAVVNGASGAVGLADD202 pKa = 4.83DD203 pKa = 4.79AVTGAGVQAAQDD215 pKa = 3.68MSTLRR220 pKa = 11.84DD221 pKa = 3.39GLAAAVTPIANATVATAAAARR242 pKa = 11.84QALTQGIEE250 pKa = 4.01DD251 pKa = 3.91TTEE254 pKa = 3.42QFAPVLAPLRR264 pKa = 11.84PGSASEE270 pKa = 4.06ALAPAVAQIGDD281 pKa = 3.33WWRR284 pKa = 11.84QSQEE288 pKa = 3.69RR289 pKa = 11.84ARR291 pKa = 11.84SSLNDD296 pKa = 3.44FLTPALAGVANVVSPQKK313 pKa = 10.53RR314 pKa = 11.84EE315 pKa = 3.34EE316 pKa = 4.13CMQKK320 pKa = 8.79FTHH323 pKa = 6.93CGFCSPNFDD332 pKa = 3.17RR333 pKa = 11.84CLFCTAPFTLIDD345 pKa = 4.34ISCACPPGTFVSSDD359 pKa = 3.25GSSCIPFAVAQLEE372 pKa = 4.1KK373 pKa = 10.7LILAISQQNAALADD387 pKa = 3.93RR388 pKa = 11.84VNSLLHH394 pKa = 6.18LVGPSRR400 pKa = 11.84TCQANIPFCLRR411 pKa = 11.84CSSPATCAVCSDD423 pKa = 4.28GYY425 pKa = 11.47KK426 pKa = 10.23LDD428 pKa = 4.26AVGKK432 pKa = 7.93CICDD436 pKa = 3.5PTTAGPGCAFCIVPGSCGNPLGGTKK461 pKa = 10.58ANLKK465 pKa = 10.07ACADD469 pKa = 3.75FGKK472 pKa = 10.1RR473 pKa = 11.84RR474 pKa = 11.84QYY476 pKa = 9.96GRR478 pKa = 11.84AYY480 pKa = 10.81NFVKK484 pKa = 10.8DD485 pKa = 3.72PVTGLGRR492 pKa = 11.84CEE494 pKa = 4.99CNPQSTPHH502 pKa = 6.49CAACLFPGVCKK513 pKa = 10.59FCNAPFEE520 pKa = 4.52LVDD523 pKa = 4.62GKK525 pKa = 10.05CACPPLYY532 pKa = 10.25GPPVMDD538 pKa = 5.34EE539 pKa = 4.22DD540 pKa = 4.0TCLPIFLGQQVYY552 pKa = 7.79ITGPTGPTGFGPTGPTGIAIGATGFTGSTAYY583 pKa = 10.32GVTGFTGFTGFTGFTGFTGPTGFTGFTGDD612 pKa = 3.06TGFTGFTGDD621 pKa = 3.06TGFTGFTGDD630 pKa = 3.06TGFTGFTGDD639 pKa = 3.22TGFTGTTGDD648 pKa = 3.25TGFTGFTGDD657 pKa = 3.46TGFTGNTGDD666 pKa = 3.72TGFTGATGFTGDD678 pKa = 3.64TGFTGDD684 pKa = 3.77TGFTGNTGDD693 pKa = 3.72TGFTGATGFTGDD705 pKa = 3.43TGTTGFTGTTGNTGDD720 pKa = 3.7TGFTGDD726 pKa = 3.32TGFTGFTGATGDD738 pKa = 3.57TGFTGNTGDD747 pKa = 3.39TGFTGFTGNTGFTGFTGDD765 pKa = 3.31TGFTGDD771 pKa = 3.72TGFTGATGFTGDD783 pKa = 3.31TGFTGFTGSTGFTGFTGDD801 pKa = 3.06TGFTGFTGFTGATGNTGDD819 pKa = 3.62TGFTGFTGDD828 pKa = 3.06TGFTGFTGDD837 pKa = 3.06TGFTGFTGFTGDD849 pKa = 3.46TGFTGNTGDD858 pKa = 3.72TGFTGATGFSGSTGFTGDD876 pKa = 3.18TGFAGATGFTGFTGDD891 pKa = 3.42TGFTGATGVTGFTGATGYY909 pKa = 10.91SGDD912 pKa = 3.83TGFTGATGFSGFTGFTGGTGFTGFTGDD939 pKa = 3.03TGFTGFPGFTGATGFTGFTGATGFTGDD966 pKa = 3.31TGFTGFTGNTGFTGATGFTGFTGFTGDD993 pKa = 3.42TGFTGATGFTGFTGATGFTGSTGFTGDD1020 pKa = 2.93TGFTGVSGFTGDD1032 pKa = 3.32TGFTGATGFTGFTGATGFTGDD1053 pKa = 4.0TGISGATGFTGFTGFTGATGFTGTTGFTGATGLTGDD1089 pKa = 4.18SGFTGTTGFTGFTGDD1104 pKa = 3.06TGFTGFTGFTGFTGATGFTGDD1125 pKa = 4.09TGNTGATGFTGFTGATGFTGDD1146 pKa = 3.31TGFTGFTGATGFTGATGFTGDD1167 pKa = 3.31TGFTGFTGFTGFTGDD1182 pKa = 3.42TGFTGATGFTGFTGATGITGDD1203 pKa = 3.89TGITGATGFTGATGYY1218 pKa = 9.12TGFTGNTGSTGSTGATGFTGATGYY1242 pKa = 9.24TGTTGFTGFTGDD1254 pKa = 3.46TGFTGNTGDD1263 pKa = 3.43TGFTGVTGFTGATGFTGSTGDD1284 pKa = 3.36TGFTGATGFTGFTGATGFTGSTGFTGTTGFTGATGFTGFTGFTGTTGATGFTGFTGATGFTGFTGFTGATGFTGATGSTGFTGFTGDD1371 pKa = 3.5TGITGATGFTGDD1383 pKa = 3.55TGFTGTTGFTGFTGDD1398 pKa = 3.42TGFTGATGFTGTTGFTGATGFTGFTGDD1425 pKa = 3.21TGITGGTGFTGSTGFTGNTGSTGATGITGFTGATGFTGDD1464 pKa = 3.76TGFTGATGFTGFTGDD1479 pKa = 3.42TGFTGATGSTGFTGATGFTGFTGATGFTGFTGGTGYY1515 pKa = 9.2TGTTGFTGATGFTGFTGDD1533 pKa = 3.5TGITGATGFTGSTGFTGTTGFTGFTGDD1560 pKa = 3.23TGFTGSTGATGFTGATGFTGGTGFTGFTGSTGFTGAIGFTGATGFTGFTGDD1611 pKa = 3.23TGFTGSTGFTGFTGDD1626 pKa = 3.42TGFTGATGFTGATGNTGFTGATGFTGTTGFTGFTGDD1662 pKa = 3.42TGFTGATGSTGFTGSTGFSGATGFSGATGFTGFTGATGFTGDD1704 pKa = 3.39TGATGFSGATGFTGSTGGTGATGYY1728 pKa = 10.76SGFTGATGFTGDD1740 pKa = 3.82SGFTGTTGATGYY1752 pKa = 8.98TGSTGFTGATGFTGFTGATGYY1773 pKa = 9.01TGATGFTGFTGDD1785 pKa = 3.06TGFTGFTGNTGFTGATGFTGSTGATGFTGATGLTGFTGATGSTGTSGSTGFTGATGFTGVTGATGFTGSTGFTGSTGFTGSTGFTGATGSTGATGDD1881 pKa = 3.43TGFTGNTGFTGNTGFTGGTGATGYY1905 pKa = 8.99TGATGFTGFTGTTGDD1920 pKa = 3.42TGITGSTGYY1929 pKa = 8.97TGSTGATGFSGATGFTGFTGDD1950 pKa = 3.42TGFTGATGFTGSTGVTGFTGGTGYY1974 pKa = 9.11TGFTGSTGATGFTGVTGFTGDD1995 pKa = 3.06TGATGFSGATGATGVTGSTGFTGSTGFTGATGFTGDD2031 pKa = 3.7TGATGLTGASGFTGNTGFTGATGFTGFTGATGFTGFTGFTGATGFTGNTGATGASGFTGSTGGTGFTGATGFTGSTGFTGFTGDD2115 pKa = 3.64TGFSGATGFTGSTGGTGTTGATGFTGSTGGTGSSGATGATGGSGATGFTGGTGATGLTGNTGATGSTGGTGFTGTTGGTGATGDD2199 pKa = 3.63TGSTGGTGITGVTGGTGALGSTGATGFTGDD2229 pKa = 3.81TGITGATGFSGATGATGFSGPSGVTGITGATGFTGSSGDD2268 pKa = 3.53TGVTGATGSRR2278 pKa = 11.84GFIGATGFTGAIGTTGSTGFTGGTGATGGTGGTGFTGATGSTGTTGGTGGTGGTGASGSTGNTGASGFTGSTGGTGATGFTGQTGGTGSTGTTGATGSTGTTGSSGITGSTGSTGSSGLQGFTGATGATGNSGTTGATGLTGASGATGFTGSTGGSGATGSTGFSGATGTTGATGFSGGSGATGSTGFSGATGGTGSTGGTGSTGATGGTGFSGATGFTGTTGGTGFTGFSGATGGSGFTGATGAAATGATGVTGNTGPTGGTGPSGSTGAVGTSGATGSTGVTGTTGTSGPTGFSGGTGSTGPGITGATGVTGPTGPTGPTGFTGGTGGTGGTGFAATGATGPTGPTGAQGVTGGAGG2636 pKa = 2.87
Molecular weight: 244.36 kDa
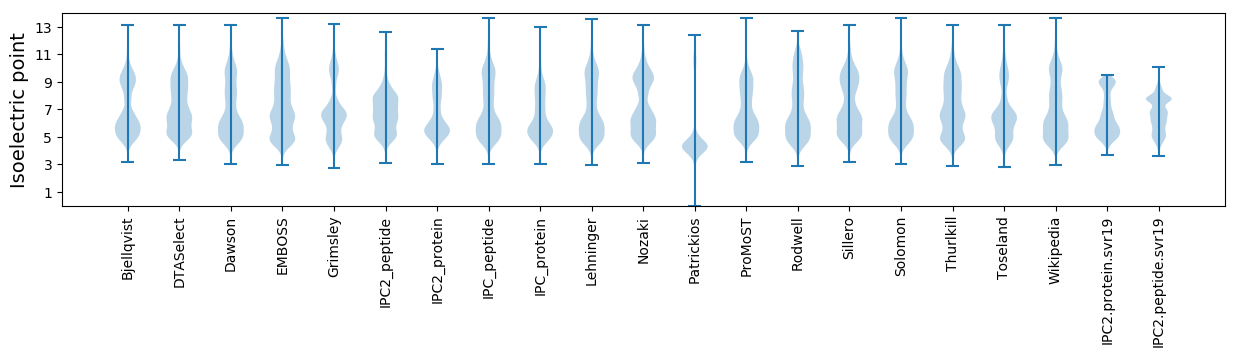
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I0Z022|I0Z022_COCSC DnaJ-domain-containing protein OS=Coccomyxa subellipsoidea (strain C-169) OX=574566 GN=COCSUDRAFT_53179 PE=4 SV=1
MM1 pKa = 7.59SLPLAPPATRR11 pKa = 11.84PKK13 pKa = 9.98HH14 pKa = 5.68SKK16 pKa = 10.24RR17 pKa = 11.84SPLSAQTRR25 pKa = 11.84QQRR28 pKa = 11.84PRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84ASAVMMSALTQALRR48 pKa = 11.84RR49 pKa = 11.84SRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84TAGRR57 pKa = 11.84AA58 pKa = 3.13
MM1 pKa = 7.59SLPLAPPATRR11 pKa = 11.84PKK13 pKa = 9.98HH14 pKa = 5.68SKK16 pKa = 10.24RR17 pKa = 11.84SPLSAQTRR25 pKa = 11.84QQRR28 pKa = 11.84PRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84ASAVMMSALTQALRR48 pKa = 11.84RR49 pKa = 11.84SRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84TAGRR57 pKa = 11.84AA58 pKa = 3.13
Molecular weight: 6.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4166057 |
31 |
15797 |
425.2 |
46.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.249 ± 0.063 | 1.525 ± 0.012 |
5.111 ± 0.019 | 6.122 ± 0.027 |
3.241 ± 0.02 | 7.899 ± 0.042 |
2.278 ± 0.011 | 3.633 ± 0.022 |
4.17 ± 0.023 | 9.739 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.24 ± 0.01 | 2.77 ± 0.018 |
5.941 ± 0.031 | 4.575 ± 0.021 |
6.253 ± 0.026 | 7.187 ± 0.024 |
4.961 ± 0.041 | 6.525 ± 0.022 |
1.356 ± 0.011 | 2.226 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
