
Jaapia argillacea MUCL 33604
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Jaapiales; Jaapiaceae; Jaapia; Jaapia argillacea
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
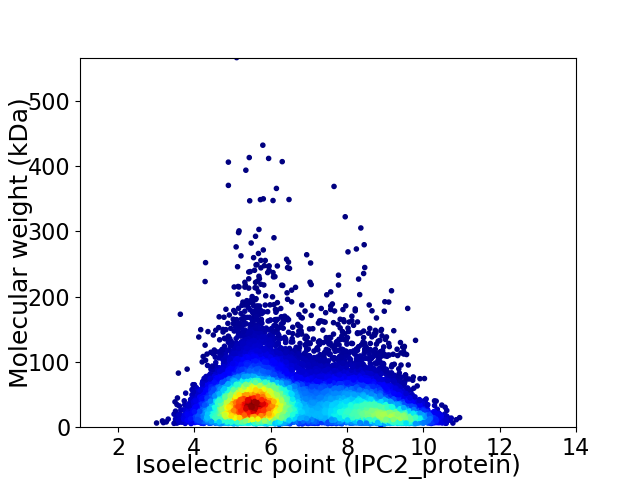
Virtual 2D-PAGE plot for 16341 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

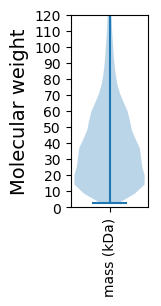
Protein with the lowest isoelectric point:
>tr|A0A067Q6T3|A0A067Q6T3_9AGAM Uncharacterized protein OS=Jaapia argillacea MUCL 33604 OX=933084 GN=JAAARDRAFT_655672 PE=4 SV=1
MM1 pKa = 6.97LHH3 pKa = 7.31KK4 pKa = 9.59YY5 pKa = 7.82TNHH8 pKa = 6.37GDD10 pKa = 3.85LLSSASHH17 pKa = 6.69EE18 pKa = 4.14GMEE21 pKa = 4.32MDD23 pKa = 3.74WVPVQSEE30 pKa = 4.64EE31 pKa = 4.27PVWLYY36 pKa = 11.16GSQDD40 pKa = 2.69IYY42 pKa = 11.53QNDD45 pKa = 3.82VQVHH49 pKa = 4.66MGEE52 pKa = 4.12HH53 pKa = 6.27LEE55 pKa = 3.98NRR57 pKa = 11.84GVFTQDD63 pKa = 2.53VFGAPAIQGTSFVDD77 pKa = 2.97KK78 pKa = 10.25DD79 pKa = 3.71EE80 pKa = 5.07RR81 pKa = 11.84MSPPPSSPSYY91 pKa = 10.44HH92 pKa = 6.27QDD94 pKa = 3.52APPMSYY100 pKa = 10.65PSIRR104 pKa = 11.84PPLTHH109 pKa = 6.39HH110 pKa = 7.01TNPTSPSPDD119 pKa = 3.26CSHH122 pKa = 7.59RR123 pKa = 11.84DD124 pKa = 3.72TNHH127 pKa = 5.71MALNSFEE134 pKa = 5.22DD135 pKa = 4.64LPDD138 pKa = 4.52GDD140 pKa = 4.2IAPPSSQDD148 pKa = 2.99PRR150 pKa = 11.84IAPRR154 pKa = 11.84ANYY157 pKa = 9.21VALDD161 pKa = 3.88SFADD165 pKa = 4.53LPDD168 pKa = 5.42DD169 pKa = 6.48DD170 pKa = 5.04IAPPNNDD177 pKa = 2.37ITPPGSQEE185 pKa = 3.57MQIIVPCGRR194 pKa = 11.84HH195 pKa = 4.16VQSPRR200 pKa = 11.84QDD202 pKa = 3.35MQEE205 pKa = 4.53PLPPMYY211 pKa = 9.86IDD213 pKa = 5.19GDD215 pKa = 4.05DD216 pKa = 5.26DD217 pKa = 4.49INPPSDD223 pKa = 4.7DD224 pKa = 3.94LPTYY228 pKa = 10.38IDD230 pKa = 3.98NDD232 pKa = 3.67NNIPPPSQEE241 pKa = 4.29ALVASPTHH249 pKa = 6.4YY250 pKa = 10.1IDD252 pKa = 4.79SDD254 pKa = 4.09DD255 pKa = 5.73DD256 pKa = 3.81IPPPSQEE263 pKa = 4.59APTFSPPSYY272 pKa = 10.31INKK275 pKa = 9.35DD276 pKa = 3.67DD277 pKa = 5.3NIPPPSQEE285 pKa = 4.33ALTFLPPTYY294 pKa = 10.55INDD297 pKa = 4.62DD298 pKa = 4.13DD299 pKa = 6.18DD300 pKa = 4.49ISPPSQEE307 pKa = 5.16AIILSPSIHH316 pKa = 6.7INNNDD321 pKa = 3.04ILTSHH326 pKa = 7.19
MM1 pKa = 6.97LHH3 pKa = 7.31KK4 pKa = 9.59YY5 pKa = 7.82TNHH8 pKa = 6.37GDD10 pKa = 3.85LLSSASHH17 pKa = 6.69EE18 pKa = 4.14GMEE21 pKa = 4.32MDD23 pKa = 3.74WVPVQSEE30 pKa = 4.64EE31 pKa = 4.27PVWLYY36 pKa = 11.16GSQDD40 pKa = 2.69IYY42 pKa = 11.53QNDD45 pKa = 3.82VQVHH49 pKa = 4.66MGEE52 pKa = 4.12HH53 pKa = 6.27LEE55 pKa = 3.98NRR57 pKa = 11.84GVFTQDD63 pKa = 2.53VFGAPAIQGTSFVDD77 pKa = 2.97KK78 pKa = 10.25DD79 pKa = 3.71EE80 pKa = 5.07RR81 pKa = 11.84MSPPPSSPSYY91 pKa = 10.44HH92 pKa = 6.27QDD94 pKa = 3.52APPMSYY100 pKa = 10.65PSIRR104 pKa = 11.84PPLTHH109 pKa = 6.39HH110 pKa = 7.01TNPTSPSPDD119 pKa = 3.26CSHH122 pKa = 7.59RR123 pKa = 11.84DD124 pKa = 3.72TNHH127 pKa = 5.71MALNSFEE134 pKa = 5.22DD135 pKa = 4.64LPDD138 pKa = 4.52GDD140 pKa = 4.2IAPPSSQDD148 pKa = 2.99PRR150 pKa = 11.84IAPRR154 pKa = 11.84ANYY157 pKa = 9.21VALDD161 pKa = 3.88SFADD165 pKa = 4.53LPDD168 pKa = 5.42DD169 pKa = 6.48DD170 pKa = 5.04IAPPNNDD177 pKa = 2.37ITPPGSQEE185 pKa = 3.57MQIIVPCGRR194 pKa = 11.84HH195 pKa = 4.16VQSPRR200 pKa = 11.84QDD202 pKa = 3.35MQEE205 pKa = 4.53PLPPMYY211 pKa = 9.86IDD213 pKa = 5.19GDD215 pKa = 4.05DD216 pKa = 5.26DD217 pKa = 4.49INPPSDD223 pKa = 4.7DD224 pKa = 3.94LPTYY228 pKa = 10.38IDD230 pKa = 3.98NDD232 pKa = 3.67NNIPPPSQEE241 pKa = 4.29ALVASPTHH249 pKa = 6.4YY250 pKa = 10.1IDD252 pKa = 4.79SDD254 pKa = 4.09DD255 pKa = 5.73DD256 pKa = 3.81IPPPSQEE263 pKa = 4.59APTFSPPSYY272 pKa = 10.31INKK275 pKa = 9.35DD276 pKa = 3.67DD277 pKa = 5.3NIPPPSQEE285 pKa = 4.33ALTFLPPTYY294 pKa = 10.55INDD297 pKa = 4.62DD298 pKa = 4.13DD299 pKa = 6.18DD300 pKa = 4.49ISPPSQEE307 pKa = 5.16AIILSPSIHH316 pKa = 6.7INNNDD321 pKa = 3.04ILTSHH326 pKa = 7.19
Molecular weight: 35.97 kDa
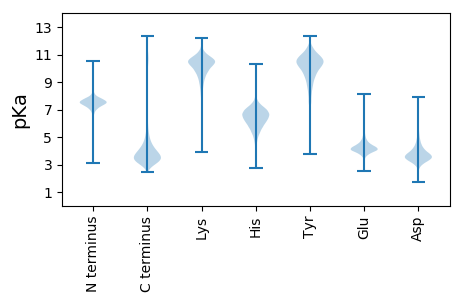
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A067QPK5|A0A067QPK5_9AGAM Uncharacterized protein OS=Jaapia argillacea MUCL 33604 OX=933084 GN=JAAARDRAFT_684114 PE=4 SV=1
MM1 pKa = 7.35CMWLRR6 pKa = 11.84GSMSMSVSLNFWGIPARR23 pKa = 11.84RR24 pKa = 11.84TSRR27 pKa = 11.84RR28 pKa = 11.84SLPTPPSAAAATTTMPVPNRR48 pKa = 11.84LLWRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84LQFYY59 pKa = 10.27SPSPTSTATSLPTTRR74 pKa = 11.84PMLLFVTPFVLRR86 pKa = 11.84TVSILPPTTTTTTTTTILTIPLPASRR112 pKa = 11.84INPSSMSIPIAASINAPRR130 pKa = 11.84ITQQ133 pKa = 3.24
MM1 pKa = 7.35CMWLRR6 pKa = 11.84GSMSMSVSLNFWGIPARR23 pKa = 11.84RR24 pKa = 11.84TSRR27 pKa = 11.84RR28 pKa = 11.84SLPTPPSAAAATTTMPVPNRR48 pKa = 11.84LLWRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84LQFYY59 pKa = 10.27SPSPTSTATSLPTTRR74 pKa = 11.84PMLLFVTPFVLRR86 pKa = 11.84TVSILPPTTTTTTTTTILTIPLPASRR112 pKa = 11.84INPSSMSIPIAASINAPRR130 pKa = 11.84ITQQ133 pKa = 3.24
Molecular weight: 14.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6436100 |
22 |
5100 |
393.9 |
43.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.739 ± 0.017 | 1.387 ± 0.009 |
5.496 ± 0.014 | 5.869 ± 0.02 |
3.784 ± 0.013 | 6.679 ± 0.018 |
2.656 ± 0.01 | 4.92 ± 0.014 |
4.415 ± 0.02 | 9.463 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.074 ± 0.007 | 3.302 ± 0.01 |
6.844 ± 0.026 | 3.642 ± 0.012 |
6.144 ± 0.017 | 9.001 ± 0.027 |
6.036 ± 0.013 | 6.427 ± 0.013 |
1.516 ± 0.007 | 2.607 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
