
Gabek Forest virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Gabek phlebovirus
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
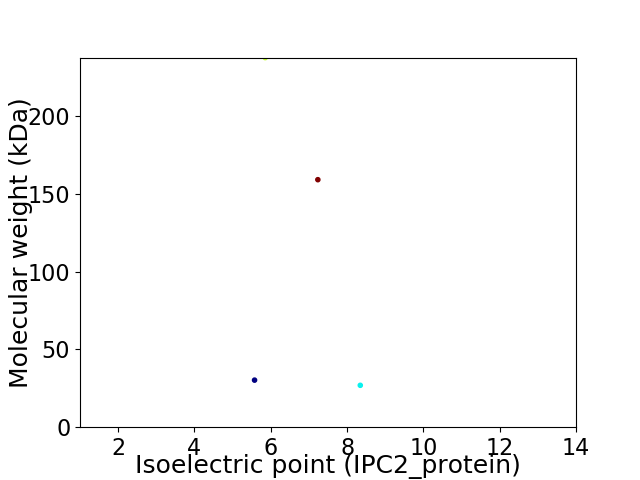
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8JX57|W8JX57_9VIRU Nonstructural protein OS=Gabek Forest virus OX=629736 PE=4 SV=1
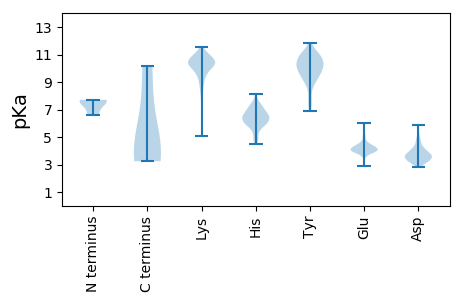
MM1 pKa = 6.63TTRR4 pKa = 11.84FLYY7 pKa = 10.55DD8 pKa = 3.18RR9 pKa = 11.84LYY11 pKa = 10.62IMQSEE16 pKa = 4.59GQKK19 pKa = 10.17LQAVFLAHH27 pKa = 6.49NNFVGDD33 pKa = 3.96DD34 pKa = 3.69VSSYY38 pKa = 11.85DD39 pKa = 3.22HH40 pKa = 6.59MEE42 pKa = 4.2VVLSKK47 pKa = 10.63YY48 pKa = 9.88KK49 pKa = 10.5QSFDD53 pKa = 3.56YY54 pKa = 10.96RR55 pKa = 11.84EE56 pKa = 4.19SLSDD60 pKa = 3.96FYY62 pKa = 11.82SRR64 pKa = 11.84GEE66 pKa = 3.87LPFRR70 pKa = 11.84WGSIMWCSRR79 pKa = 11.84VTGEE83 pKa = 4.53EE84 pKa = 4.15YY85 pKa = 10.83LSFMALMMDD94 pKa = 4.59LLKK97 pKa = 10.69LDD99 pKa = 3.61SSEE102 pKa = 4.66LKK104 pKa = 10.74GDD106 pKa = 3.21EE107 pKa = 4.31HH108 pKa = 7.29PNIRR112 pKa = 11.84EE113 pKa = 3.99ALSWPTGSPTLGFIKK128 pKa = 10.36LNCRR132 pKa = 11.84SSDD135 pKa = 2.82WSINFQKK142 pKa = 10.77SRR144 pKa = 11.84TATLLLRR151 pKa = 11.84AGGSDD156 pKa = 3.32EE157 pKa = 4.15GLEE160 pKa = 3.89EE161 pKa = 5.92AIVKK165 pKa = 6.04THH167 pKa = 6.23KK168 pKa = 10.38KK169 pKa = 10.02IIMEE173 pKa = 3.93SSIRR177 pKa = 11.84GFDD180 pKa = 3.29PKK182 pKa = 10.63FFPGLDD188 pKa = 4.19LIRR191 pKa = 11.84EE192 pKa = 4.38VACLQCVRR200 pKa = 11.84LMNASCFDD208 pKa = 4.18TVHH211 pKa = 6.04TAYY214 pKa = 9.95PSRR217 pKa = 11.84LLDD220 pKa = 3.81LLAMHH225 pKa = 7.41RR226 pKa = 11.84STYY229 pKa = 10.14TMISSKK235 pKa = 10.93LLGNRR240 pKa = 11.84KK241 pKa = 5.11WTPVQGTHH249 pKa = 6.48FDD251 pKa = 4.05EE252 pKa = 5.52PDD254 pKa = 2.99ATYY257 pKa = 11.05SFSSDD262 pKa = 3.14SEE264 pKa = 4.24
MM1 pKa = 6.63TTRR4 pKa = 11.84FLYY7 pKa = 10.55DD8 pKa = 3.18RR9 pKa = 11.84LYY11 pKa = 10.62IMQSEE16 pKa = 4.59GQKK19 pKa = 10.17LQAVFLAHH27 pKa = 6.49NNFVGDD33 pKa = 3.96DD34 pKa = 3.69VSSYY38 pKa = 11.85DD39 pKa = 3.22HH40 pKa = 6.59MEE42 pKa = 4.2VVLSKK47 pKa = 10.63YY48 pKa = 9.88KK49 pKa = 10.5QSFDD53 pKa = 3.56YY54 pKa = 10.96RR55 pKa = 11.84EE56 pKa = 4.19SLSDD60 pKa = 3.96FYY62 pKa = 11.82SRR64 pKa = 11.84GEE66 pKa = 3.87LPFRR70 pKa = 11.84WGSIMWCSRR79 pKa = 11.84VTGEE83 pKa = 4.53EE84 pKa = 4.15YY85 pKa = 10.83LSFMALMMDD94 pKa = 4.59LLKK97 pKa = 10.69LDD99 pKa = 3.61SSEE102 pKa = 4.66LKK104 pKa = 10.74GDD106 pKa = 3.21EE107 pKa = 4.31HH108 pKa = 7.29PNIRR112 pKa = 11.84EE113 pKa = 3.99ALSWPTGSPTLGFIKK128 pKa = 10.36LNCRR132 pKa = 11.84SSDD135 pKa = 2.82WSINFQKK142 pKa = 10.77SRR144 pKa = 11.84TATLLLRR151 pKa = 11.84AGGSDD156 pKa = 3.32EE157 pKa = 4.15GLEE160 pKa = 3.89EE161 pKa = 5.92AIVKK165 pKa = 6.04THH167 pKa = 6.23KK168 pKa = 10.38KK169 pKa = 10.02IIMEE173 pKa = 3.93SSIRR177 pKa = 11.84GFDD180 pKa = 3.29PKK182 pKa = 10.63FFPGLDD188 pKa = 4.19LIRR191 pKa = 11.84EE192 pKa = 4.38VACLQCVRR200 pKa = 11.84LMNASCFDD208 pKa = 4.18TVHH211 pKa = 6.04TAYY214 pKa = 9.95PSRR217 pKa = 11.84LLDD220 pKa = 3.81LLAMHH225 pKa = 7.41RR226 pKa = 11.84STYY229 pKa = 10.14TMISSKK235 pKa = 10.93LLGNRR240 pKa = 11.84KK241 pKa = 5.11WTPVQGTHH249 pKa = 6.48FDD251 pKa = 4.05EE252 pKa = 5.52PDD254 pKa = 2.99ATYY257 pKa = 11.05SFSSDD262 pKa = 3.14SEE264 pKa = 4.24
Molecular weight: 30.23 kDa
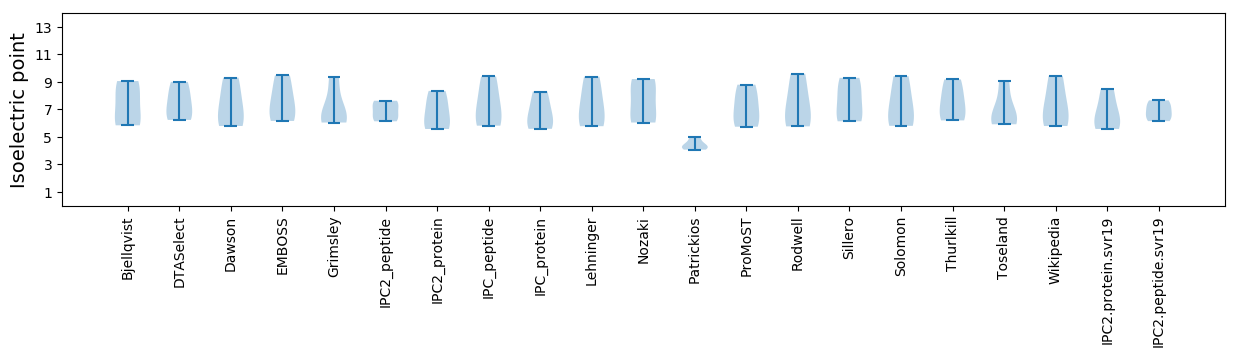
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8JDL6|W8JDL6_9VIRU Membrane glycoprotein OS=Gabek Forest virus OX=629736 PE=4 SV=1
MM1 pKa = 7.7SDD3 pKa = 3.29YY4 pKa = 11.22AQIAIEE10 pKa = 4.22FGNSEE15 pKa = 3.88VDD17 pKa = 3.18GDD19 pKa = 5.52LIEE22 pKa = 4.9AWVKK26 pKa = 9.49EE27 pKa = 3.89FEE29 pKa = 4.46YY30 pKa = 10.74QGFDD34 pKa = 3.18PRR36 pKa = 11.84IVVKK40 pKa = 10.49LVSEE44 pKa = 4.4VEE46 pKa = 4.14GWQTDD51 pKa = 3.73VKK53 pKa = 11.29KK54 pKa = 10.46MIILALTRR62 pKa = 11.84GNKK65 pKa = 7.72PEE67 pKa = 3.99KK68 pKa = 9.83MVTKK72 pKa = 9.73MSAKK76 pKa = 9.86GRR78 pKa = 11.84EE79 pKa = 4.02EE80 pKa = 3.75VAKK83 pKa = 10.47LVKK86 pKa = 10.05KK87 pKa = 10.85YY88 pKa = 10.36KK89 pKa = 10.34LKK91 pKa = 10.78SGNPGRR97 pKa = 11.84NDD99 pKa = 3.09LTLSRR104 pKa = 11.84VAAAFASWTCNAIYY118 pKa = 9.89HH119 pKa = 4.86VQYY122 pKa = 10.51YY123 pKa = 10.51LPVTGNHH130 pKa = 5.71MDD132 pKa = 5.16AISKK136 pKa = 9.78NYY138 pKa = 7.95PRR140 pKa = 11.84PMMHH144 pKa = 7.22PSFAGLIDD152 pKa = 3.45PAMRR156 pKa = 11.84DD157 pKa = 3.37RR158 pKa = 11.84QILIEE163 pKa = 4.05AHH165 pKa = 6.27SLFLIEE171 pKa = 5.02FAQKK175 pKa = 10.53INVNLRR181 pKa = 11.84GKK183 pKa = 10.12SKK185 pKa = 11.49AEE187 pKa = 3.45ICLSFDD193 pKa = 3.33QPLNAAINSNFLTSAQKK210 pKa = 10.75LKK212 pKa = 11.04ALTALGIIDD221 pKa = 4.76DD222 pKa = 4.14HH223 pKa = 6.6QKK225 pKa = 10.29PSKK228 pKa = 10.69DD229 pKa = 3.5VVNAADD235 pKa = 3.4AFRR238 pKa = 11.84RR239 pKa = 11.84II240 pKa = 3.89
MM1 pKa = 7.7SDD3 pKa = 3.29YY4 pKa = 11.22AQIAIEE10 pKa = 4.22FGNSEE15 pKa = 3.88VDD17 pKa = 3.18GDD19 pKa = 5.52LIEE22 pKa = 4.9AWVKK26 pKa = 9.49EE27 pKa = 3.89FEE29 pKa = 4.46YY30 pKa = 10.74QGFDD34 pKa = 3.18PRR36 pKa = 11.84IVVKK40 pKa = 10.49LVSEE44 pKa = 4.4VEE46 pKa = 4.14GWQTDD51 pKa = 3.73VKK53 pKa = 11.29KK54 pKa = 10.46MIILALTRR62 pKa = 11.84GNKK65 pKa = 7.72PEE67 pKa = 3.99KK68 pKa = 9.83MVTKK72 pKa = 9.73MSAKK76 pKa = 9.86GRR78 pKa = 11.84EE79 pKa = 4.02EE80 pKa = 3.75VAKK83 pKa = 10.47LVKK86 pKa = 10.05KK87 pKa = 10.85YY88 pKa = 10.36KK89 pKa = 10.34LKK91 pKa = 10.78SGNPGRR97 pKa = 11.84NDD99 pKa = 3.09LTLSRR104 pKa = 11.84VAAAFASWTCNAIYY118 pKa = 9.89HH119 pKa = 4.86VQYY122 pKa = 10.51YY123 pKa = 10.51LPVTGNHH130 pKa = 5.71MDD132 pKa = 5.16AISKK136 pKa = 9.78NYY138 pKa = 7.95PRR140 pKa = 11.84PMMHH144 pKa = 7.22PSFAGLIDD152 pKa = 3.45PAMRR156 pKa = 11.84DD157 pKa = 3.37RR158 pKa = 11.84QILIEE163 pKa = 4.05AHH165 pKa = 6.27SLFLIEE171 pKa = 5.02FAQKK175 pKa = 10.53INVNLRR181 pKa = 11.84GKK183 pKa = 10.12SKK185 pKa = 11.49AEE187 pKa = 3.45ICLSFDD193 pKa = 3.33QPLNAAINSNFLTSAQKK210 pKa = 10.75LKK212 pKa = 11.04ALTALGIIDD221 pKa = 4.76DD222 pKa = 4.14HH223 pKa = 6.6QKK225 pKa = 10.29PSKK228 pKa = 10.69DD229 pKa = 3.5VVNAADD235 pKa = 3.4AFRR238 pKa = 11.84RR239 pKa = 11.84II240 pKa = 3.89
Molecular weight: 26.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4034 |
240 |
2102 |
1008.5 |
113.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.801 ± 0.636 | 2.553 ± 0.594 |
5.156 ± 0.522 | 7.412 ± 0.494 |
4.388 ± 0.589 | 6.197 ± 0.255 |
2.305 ± 0.121 | 6.916 ± 0.56 |
6.743 ± 0.5 | 8.528 ± 0.467 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.123 ± 0.45 | 4.09 ± 0.415 |
3.867 ± 0.197 | 2.876 ± 0.302 |
5.578 ± 0.227 | 9.048 ± 0.402 |
5.751 ± 0.595 | 5.925 ± 0.243 |
1.19 ± 0.158 | 2.553 ± 0.249 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
