
bacterium J17
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
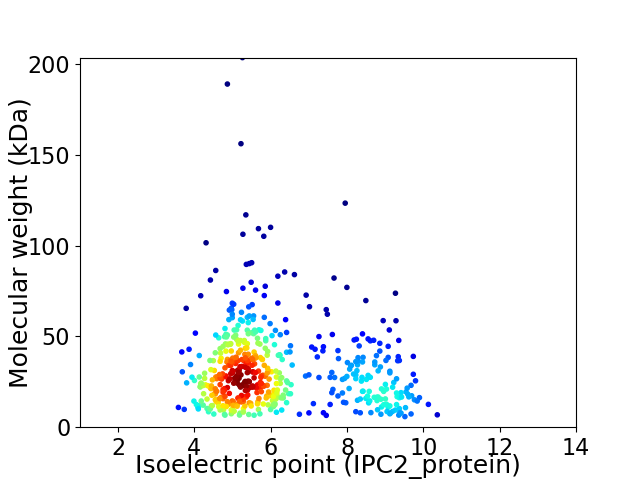
Virtual 2D-PAGE plot for 527 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A202DUX6|A0A202DUX6_9BACT Uncharacterized protein OS=bacterium J17 OX=1932695 GN=BVY02_02200 PE=4 SV=1
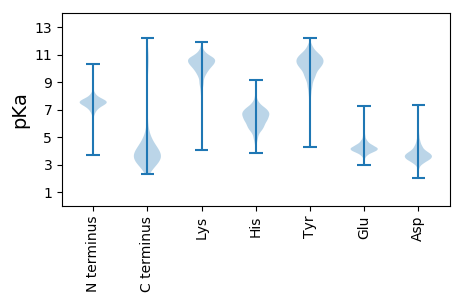
KKK2 pKa = 10.07KK3 pKa = 10.55NDDD6 pKa = 4.06AVGARR11 pKa = 11.84TDDD14 pKa = 3.35DDD16 pKa = 3.54GSGNGAVYYY25 pKa = 10.35LFLNTTGVVDDD36 pKa = 4.5YYY38 pKa = 11.81KKK40 pKa = 10.67SDDD43 pKa = 3.91EEE45 pKa = 4.35SFTAILDDD53 pKa = 3.74GQFGMSVTNTGDDD66 pKa = 3.27DDD68 pKa = 3.74DDD70 pKa = 3.97VNDDD74 pKa = 4.64GVGAYYY80 pKa = 10.17DDD82 pKa = 5.8DD83 pKa = 5.71DD84 pKa = 4.42GDDD87 pKa = 3.38RR88 pKa = 11.84GAFYYY93 pKa = 11.17LFMQTTGTVSSFQKKK108 pKa = 10.77SDDD111 pKa = 3.56AGSFTATLDDD121 pKa = 3.25TDDD124 pKa = 3.67LGVSISNAGDDD135 pKa = 3.76DDD137 pKa = 4.2DDD139 pKa = 4.42VDDD142 pKa = 4.56EE143 pKa = 4.63ASGAWNDDD151 pKa = 4.72DD152 pKa = 3.4DD153 pKa = 6.03GTNRR157 pKa = 11.84GAFYYY162 pKa = 10.58LNLDDD167 pKa = 3.84PNGNSIGTLTINSHHH182 pKa = 6.74FLSIPTFSVSNNLTLTFTTEEE203 pKa = 3.51AIPANGDDD211 pKa = 2.54DDD213 pKa = 4.31IFPIGFDDD221 pKa = 3.36TSIGSSDDD229 pKa = 3.48SEEE232 pKa = 4.65GGDDD236 pKa = 3.57GTLTTSINGHHH247 pKa = 5.95MTINTGSATSASTQFALTILNLVNPVTVGTYYY279 pKa = 8.6TFLIEEE285 pKa = 4.24QDDD288 pKa = 3.47EE289 pKa = 4.38NAVIDDD295 pKa = 3.77GVGNAVDDD303 pKa = 4.12VSDDD307 pKa = 3.5YYY309 pKa = 11.11YYY311 pKa = 10.17EE312 pKa = 3.86SKKK315 pKa = 11.6EEE317 pKa = 4.82DD318 pKa = 4.07NNSYYY323 pKa = 10.97KKK325 pKa = 10.75DD326 pKa = 3.35DDD328 pKa = 3.8SLRR331 pKa = 11.84EEE333 pKa = 4.53ITAANAVGSAVTITFDDD350 pKa = 2.99TSCYYY355 pKa = 11.01STCVIAPTSTLPSLTSNYYY374 pKa = 9.54NIVGSSFLSGISRR387 pKa = 11.84HHH389 pKa = 5.66IFALVSCARR398 pKa = 11.84ICV
KKK2 pKa = 10.07KK3 pKa = 10.55NDDD6 pKa = 4.06AVGARR11 pKa = 11.84TDDD14 pKa = 3.35DDD16 pKa = 3.54GSGNGAVYYY25 pKa = 10.35LFLNTTGVVDDD36 pKa = 4.5YYY38 pKa = 11.81KKK40 pKa = 10.67SDDD43 pKa = 3.91EEE45 pKa = 4.35SFTAILDDD53 pKa = 3.74GQFGMSVTNTGDDD66 pKa = 3.27DDD68 pKa = 3.74DDD70 pKa = 3.97VNDDD74 pKa = 4.64GVGAYYY80 pKa = 10.17DDD82 pKa = 5.8DD83 pKa = 5.71DD84 pKa = 4.42GDDD87 pKa = 3.38RR88 pKa = 11.84GAFYYY93 pKa = 11.17LFMQTTGTVSSFQKKK108 pKa = 10.77SDDD111 pKa = 3.56AGSFTATLDDD121 pKa = 3.25TDDD124 pKa = 3.67LGVSISNAGDDD135 pKa = 3.76DDD137 pKa = 4.2DDD139 pKa = 4.42VDDD142 pKa = 4.56EE143 pKa = 4.63ASGAWNDDD151 pKa = 4.72DD152 pKa = 3.4DD153 pKa = 6.03GTNRR157 pKa = 11.84GAFYYY162 pKa = 10.58LNLDDD167 pKa = 3.84PNGNSIGTLTINSHHH182 pKa = 6.74FLSIPTFSVSNNLTLTFTTEEE203 pKa = 3.51AIPANGDDD211 pKa = 2.54DDD213 pKa = 4.31IFPIGFDDD221 pKa = 3.36TSIGSSDDD229 pKa = 3.48SEEE232 pKa = 4.65GGDDD236 pKa = 3.57GTLTTSINGHHH247 pKa = 5.95MTINTGSATSASTQFALTILNLVNPVTVGTYYY279 pKa = 8.6TFLIEEE285 pKa = 4.24QDDD288 pKa = 3.47EE289 pKa = 4.38NAVIDDD295 pKa = 3.77GVGNAVDDD303 pKa = 4.12VSDDD307 pKa = 3.5YYY309 pKa = 11.11YYY311 pKa = 10.17EE312 pKa = 3.86SKKK315 pKa = 11.6EEE317 pKa = 4.82DD318 pKa = 4.07NNSYYY323 pKa = 10.97KKK325 pKa = 10.75DD326 pKa = 3.35DDD328 pKa = 3.8SLRR331 pKa = 11.84EEE333 pKa = 4.53ITAANAVGSAVTITFDDD350 pKa = 2.99TSCYYY355 pKa = 11.01STCVIAPTSTLPSLTSNYYY374 pKa = 9.54NIVGSSFLSGISRR387 pKa = 11.84HHH389 pKa = 5.66IFALVSCARR398 pKa = 11.84ICV
Molecular weight: 41.47 kDa
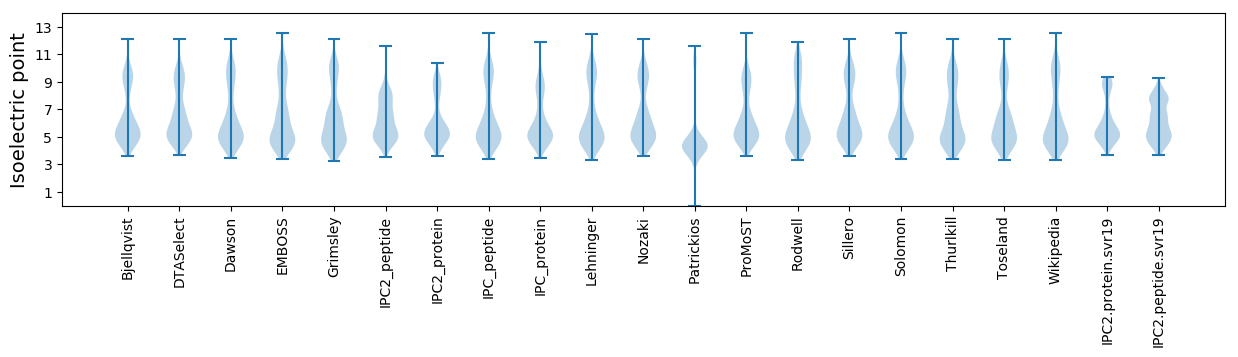
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A202DVV5|A0A202DVV5_9BACT Aminomethyltransferase OS=bacterium J17 OX=1932695 GN=gcvT PE=3 SV=1
MM1 pKa = 7.36SRR3 pKa = 11.84HH4 pKa = 4.91EE5 pKa = 4.56SNRR8 pKa = 11.84NRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 10.19NGGARR18 pKa = 11.84LSPTQRR24 pKa = 11.84SAHH27 pKa = 6.15LPLLTRR33 pKa = 11.84QKK35 pKa = 10.67EE36 pKa = 4.04LRR38 pKa = 11.84DD39 pKa = 3.61NEE41 pKa = 4.5LADD44 pKa = 4.69DD45 pKa = 4.26PLNRR49 pKa = 11.84IIAAEE54 pKa = 4.06KK55 pKa = 10.02ARR57 pKa = 11.84AGRR60 pKa = 11.84EE61 pKa = 3.81STINILMIFSSVGALVVWLGFFVLFQQIINLNWGEE96 pKa = 4.1GGQLMSRR103 pKa = 11.84RR104 pKa = 11.84VLDD107 pKa = 3.95GSSIILAQKK116 pKa = 7.57QHH118 pKa = 6.6PAAGGLSASNSLKK131 pKa = 9.92EE132 pKa = 4.0SGYY135 pKa = 8.54MRR137 pKa = 11.84GIVGEE142 pKa = 4.25VGTKK146 pKa = 7.99QVYY149 pKa = 8.74YY150 pKa = 9.93QAPPPQCLSHH160 pKa = 7.95LVMPII165 pKa = 2.98
MM1 pKa = 7.36SRR3 pKa = 11.84HH4 pKa = 4.91EE5 pKa = 4.56SNRR8 pKa = 11.84NRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 10.19NGGARR18 pKa = 11.84LSPTQRR24 pKa = 11.84SAHH27 pKa = 6.15LPLLTRR33 pKa = 11.84QKK35 pKa = 10.67EE36 pKa = 4.04LRR38 pKa = 11.84DD39 pKa = 3.61NEE41 pKa = 4.5LADD44 pKa = 4.69DD45 pKa = 4.26PLNRR49 pKa = 11.84IIAAEE54 pKa = 4.06KK55 pKa = 10.02ARR57 pKa = 11.84AGRR60 pKa = 11.84EE61 pKa = 3.81STINILMIFSSVGALVVWLGFFVLFQQIINLNWGEE96 pKa = 4.1GGQLMSRR103 pKa = 11.84RR104 pKa = 11.84VLDD107 pKa = 3.95GSSIILAQKK116 pKa = 7.57QHH118 pKa = 6.6PAAGGLSASNSLKK131 pKa = 9.92EE132 pKa = 4.0SGYY135 pKa = 8.54MRR137 pKa = 11.84GIVGEE142 pKa = 4.25VGTKK146 pKa = 7.99QVYY149 pKa = 8.74YY150 pKa = 9.93QAPPPQCLSHH160 pKa = 7.95LVMPII165 pKa = 2.98
Molecular weight: 18.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
156075 |
50 |
1856 |
296.2 |
32.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.427 ± 0.105 | 1.007 ± 0.044 |
5.365 ± 0.088 | 7.311 ± 0.116 |
4.55 ± 0.086 | 7.094 ± 0.099 |
1.674 ± 0.047 | 6.023 ± 0.071 |
5.56 ± 0.112 | 10.813 ± 0.133 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.858 ± 0.04 | 3.657 ± 0.078 |
4.061 ± 0.055 | 3.299 ± 0.057 |
5.429 ± 0.092 | 8.367 ± 0.109 |
4.564 ± 0.075 | 7.383 ± 0.121 |
0.948 ± 0.033 | 2.611 ± 0.066 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
