
endosymbiont of Galathealinum brachiosum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; sulfur-oxidizing symbionts
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
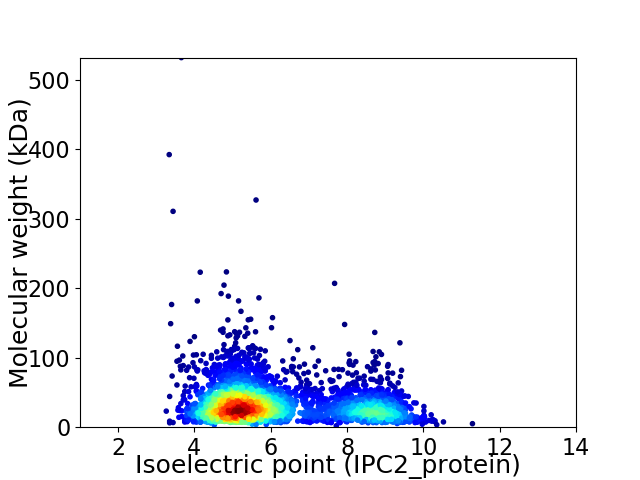
Virtual 2D-PAGE plot for 3414 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A370DC79|A0A370DC79_9GAMM Uncharacterized protein OS=endosymbiont of Galathealinum brachiosum OX=2200906 GN=DIZ80_09485 PE=4 SV=1
MM1 pKa = 7.48NIKK4 pKa = 10.61NIFSQVLFIFTVFVLVSCGGGSGDD28 pKa = 4.9AVTTTTGQFIDD39 pKa = 5.29DD40 pKa = 4.16PVQGLDD46 pKa = 3.59YY47 pKa = 10.26SCSSGTTGITNINGEE62 pKa = 4.45YY63 pKa = 9.19TCNTGDD69 pKa = 4.78DD70 pKa = 3.93VTFSIGPVTLGTLSAQSSFITPYY93 pKa = 11.22SLFPNDD99 pKa = 2.84TQAAVNLARR108 pKa = 11.84LLQSIDD114 pKa = 3.42SDD116 pKa = 4.38SNPNNNVITLSSSQLSLLPTNTDD139 pKa = 3.48FTDD142 pKa = 3.11INFVANIEE150 pKa = 4.23TALGITLIDD159 pKa = 3.46AMTAMSHH166 pKa = 6.78LYY168 pKa = 10.29DD169 pKa = 5.64AIILNGGNVPDD180 pKa = 5.06GGHH183 pKa = 6.88LPIASAGMDD192 pKa = 3.46LSVVVQNAIALDD204 pKa = 4.02GSGSYY209 pKa = 10.7DD210 pKa = 3.6ADD212 pKa = 3.84GDD214 pKa = 4.29SLTYY218 pKa = 8.83TWSIASKK225 pKa = 10.18PPGSVSFLFTGMNTSAPILTPDD247 pKa = 3.22VVGDD251 pKa = 3.72YY252 pKa = 10.2VAQLIVNDD260 pKa = 4.23GLVNSAADD268 pKa = 3.66TVTITSTVSGNVSDD282 pKa = 5.03SPPSLAWIKK291 pKa = 8.66EE292 pKa = 3.87VRR294 pKa = 11.84VEE296 pKa = 4.08TSGGQDD302 pKa = 2.75GGIAFDD308 pKa = 4.47SNNNLYY314 pKa = 9.17MAAEE318 pKa = 4.08SLIDD322 pKa = 4.64FNGDD326 pKa = 3.11LNAGYY331 pKa = 10.57QDD333 pKa = 3.44ILVMKK338 pKa = 9.56FDD340 pKa = 4.38AAGDD344 pKa = 4.04MLWSDD349 pKa = 3.59QTGTANTDD357 pKa = 3.48LVDD360 pKa = 6.27DD361 pKa = 4.02IAFDD365 pKa = 3.41KK366 pKa = 10.78SANVLYY372 pKa = 9.61VTGSVSLSLHH382 pKa = 5.34GQDD385 pKa = 4.71VIGDD389 pKa = 3.53RR390 pKa = 11.84DD391 pKa = 3.57IYY393 pKa = 10.43IISYY397 pKa = 9.7DD398 pKa = 3.56AEE400 pKa = 4.55GNRR403 pKa = 11.84LWTIMDD409 pKa = 3.78GTAQEE414 pKa = 4.07EE415 pKa = 4.38RR416 pKa = 11.84PYY418 pKa = 11.01DD419 pKa = 3.43IALDD423 pKa = 3.55SSGNIFIAGVTHH435 pKa = 7.18GNFDD439 pKa = 4.08GNTTPTGRR447 pKa = 11.84DD448 pKa = 2.73IFVAKK453 pKa = 10.4YY454 pKa = 9.78NNLGAKK460 pKa = 8.98IWTRR464 pKa = 11.84QLQGDD469 pKa = 3.23NWEE472 pKa = 4.15RR473 pKa = 11.84AEE475 pKa = 4.73GIAVDD480 pKa = 4.11SSDD483 pKa = 3.09NVYY486 pKa = 8.62ITGYY490 pKa = 8.8TGSDD494 pKa = 3.27EE495 pKa = 5.18LGGEE499 pKa = 4.33VTNGSINSFIIKK511 pKa = 9.83YY512 pKa = 10.32SNDD515 pKa = 2.93GSRR518 pKa = 11.84LWTRR522 pKa = 11.84LISSANSEE530 pKa = 4.15YY531 pKa = 10.87GYY533 pKa = 11.52GVATDD538 pKa = 3.67SNNNVFVTGMTSGDD552 pKa = 3.33LDD554 pKa = 4.02GNTNRR559 pKa = 11.84GGYY562 pKa = 9.41DD563 pKa = 3.15QFTAKK568 pKa = 10.71YY569 pKa = 7.19NTNGALIWTTLTGGTLTDD587 pKa = 3.54TANDD591 pKa = 3.27IVVDD595 pKa = 3.77SFGRR599 pKa = 11.84IYY601 pKa = 9.9ITGQIGKK608 pKa = 9.2SEE610 pKa = 4.19TGDD613 pKa = 3.01IMDD616 pKa = 4.15YY617 pKa = 10.51VLTMFDD623 pKa = 3.49TDD625 pKa = 4.36GEE627 pKa = 4.66EE628 pKa = 3.85VWTVQNTPEE637 pKa = 3.99YY638 pKa = 11.1GRR640 pKa = 11.84IRR642 pKa = 11.84DD643 pKa = 3.78DD644 pKa = 3.61RR645 pKa = 11.84GKK647 pKa = 10.09HH648 pKa = 5.33LALDD652 pKa = 3.77SAEE655 pKa = 4.09NIYY658 pKa = 11.28VNGTTYY664 pKa = 11.17GSFFSTGDD672 pKa = 3.48DD673 pKa = 3.24ASNKK677 pKa = 9.24VFVARR682 pKa = 11.84YY683 pKa = 7.69NKK685 pKa = 9.98SVAGGPYY692 pKa = 10.02SVGGYY697 pKa = 10.3VNDD700 pKa = 5.15AIGSFTLTNNGGDD713 pKa = 3.73DD714 pKa = 4.53LFILEE719 pKa = 5.11DD720 pKa = 3.91GVFTFDD726 pKa = 3.72TLLNDD731 pKa = 3.87GASYY735 pKa = 10.77NVEE738 pKa = 4.04ILDD741 pKa = 5.38GSDD744 pKa = 2.69TSQGHH749 pKa = 5.87DD750 pKa = 3.74CEE752 pKa = 4.46VLSGGSNGDD761 pKa = 2.97GSGTINGADD770 pKa = 3.32VTDD773 pKa = 4.27IEE775 pKa = 4.85VSCVGII781 pKa = 3.99
MM1 pKa = 7.48NIKK4 pKa = 10.61NIFSQVLFIFTVFVLVSCGGGSGDD28 pKa = 4.9AVTTTTGQFIDD39 pKa = 5.29DD40 pKa = 4.16PVQGLDD46 pKa = 3.59YY47 pKa = 10.26SCSSGTTGITNINGEE62 pKa = 4.45YY63 pKa = 9.19TCNTGDD69 pKa = 4.78DD70 pKa = 3.93VTFSIGPVTLGTLSAQSSFITPYY93 pKa = 11.22SLFPNDD99 pKa = 2.84TQAAVNLARR108 pKa = 11.84LLQSIDD114 pKa = 3.42SDD116 pKa = 4.38SNPNNNVITLSSSQLSLLPTNTDD139 pKa = 3.48FTDD142 pKa = 3.11INFVANIEE150 pKa = 4.23TALGITLIDD159 pKa = 3.46AMTAMSHH166 pKa = 6.78LYY168 pKa = 10.29DD169 pKa = 5.64AIILNGGNVPDD180 pKa = 5.06GGHH183 pKa = 6.88LPIASAGMDD192 pKa = 3.46LSVVVQNAIALDD204 pKa = 4.02GSGSYY209 pKa = 10.7DD210 pKa = 3.6ADD212 pKa = 3.84GDD214 pKa = 4.29SLTYY218 pKa = 8.83TWSIASKK225 pKa = 10.18PPGSVSFLFTGMNTSAPILTPDD247 pKa = 3.22VVGDD251 pKa = 3.72YY252 pKa = 10.2VAQLIVNDD260 pKa = 4.23GLVNSAADD268 pKa = 3.66TVTITSTVSGNVSDD282 pKa = 5.03SPPSLAWIKK291 pKa = 8.66EE292 pKa = 3.87VRR294 pKa = 11.84VEE296 pKa = 4.08TSGGQDD302 pKa = 2.75GGIAFDD308 pKa = 4.47SNNNLYY314 pKa = 9.17MAAEE318 pKa = 4.08SLIDD322 pKa = 4.64FNGDD326 pKa = 3.11LNAGYY331 pKa = 10.57QDD333 pKa = 3.44ILVMKK338 pKa = 9.56FDD340 pKa = 4.38AAGDD344 pKa = 4.04MLWSDD349 pKa = 3.59QTGTANTDD357 pKa = 3.48LVDD360 pKa = 6.27DD361 pKa = 4.02IAFDD365 pKa = 3.41KK366 pKa = 10.78SANVLYY372 pKa = 9.61VTGSVSLSLHH382 pKa = 5.34GQDD385 pKa = 4.71VIGDD389 pKa = 3.53RR390 pKa = 11.84DD391 pKa = 3.57IYY393 pKa = 10.43IISYY397 pKa = 9.7DD398 pKa = 3.56AEE400 pKa = 4.55GNRR403 pKa = 11.84LWTIMDD409 pKa = 3.78GTAQEE414 pKa = 4.07EE415 pKa = 4.38RR416 pKa = 11.84PYY418 pKa = 11.01DD419 pKa = 3.43IALDD423 pKa = 3.55SSGNIFIAGVTHH435 pKa = 7.18GNFDD439 pKa = 4.08GNTTPTGRR447 pKa = 11.84DD448 pKa = 2.73IFVAKK453 pKa = 10.4YY454 pKa = 9.78NNLGAKK460 pKa = 8.98IWTRR464 pKa = 11.84QLQGDD469 pKa = 3.23NWEE472 pKa = 4.15RR473 pKa = 11.84AEE475 pKa = 4.73GIAVDD480 pKa = 4.11SSDD483 pKa = 3.09NVYY486 pKa = 8.62ITGYY490 pKa = 8.8TGSDD494 pKa = 3.27EE495 pKa = 5.18LGGEE499 pKa = 4.33VTNGSINSFIIKK511 pKa = 9.83YY512 pKa = 10.32SNDD515 pKa = 2.93GSRR518 pKa = 11.84LWTRR522 pKa = 11.84LISSANSEE530 pKa = 4.15YY531 pKa = 10.87GYY533 pKa = 11.52GVATDD538 pKa = 3.67SNNNVFVTGMTSGDD552 pKa = 3.33LDD554 pKa = 4.02GNTNRR559 pKa = 11.84GGYY562 pKa = 9.41DD563 pKa = 3.15QFTAKK568 pKa = 10.71YY569 pKa = 7.19NTNGALIWTTLTGGTLTDD587 pKa = 3.54TANDD591 pKa = 3.27IVVDD595 pKa = 3.77SFGRR599 pKa = 11.84IYY601 pKa = 9.9ITGQIGKK608 pKa = 9.2SEE610 pKa = 4.19TGDD613 pKa = 3.01IMDD616 pKa = 4.15YY617 pKa = 10.51VLTMFDD623 pKa = 3.49TDD625 pKa = 4.36GEE627 pKa = 4.66EE628 pKa = 3.85VWTVQNTPEE637 pKa = 3.99YY638 pKa = 11.1GRR640 pKa = 11.84IRR642 pKa = 11.84DD643 pKa = 3.78DD644 pKa = 3.61RR645 pKa = 11.84GKK647 pKa = 10.09HH648 pKa = 5.33LALDD652 pKa = 3.77SAEE655 pKa = 4.09NIYY658 pKa = 11.28VNGTTYY664 pKa = 11.17GSFFSTGDD672 pKa = 3.48DD673 pKa = 3.24ASNKK677 pKa = 9.24VFVARR682 pKa = 11.84YY683 pKa = 7.69NKK685 pKa = 9.98SVAGGPYY692 pKa = 10.02SVGGYY697 pKa = 10.3VNDD700 pKa = 5.15AIGSFTLTNNGGDD713 pKa = 3.73DD714 pKa = 4.53LFILEE719 pKa = 5.11DD720 pKa = 3.91GVFTFDD726 pKa = 3.72TLLNDD731 pKa = 3.87GASYY735 pKa = 10.77NVEE738 pKa = 4.04ILDD741 pKa = 5.38GSDD744 pKa = 2.69TSQGHH749 pKa = 5.87DD750 pKa = 3.74CEE752 pKa = 4.46VLSGGSNGDD761 pKa = 2.97GSGTINGADD770 pKa = 3.32VTDD773 pKa = 4.27IEE775 pKa = 4.85VSCVGII781 pKa = 3.99
Molecular weight: 82.49 kDa
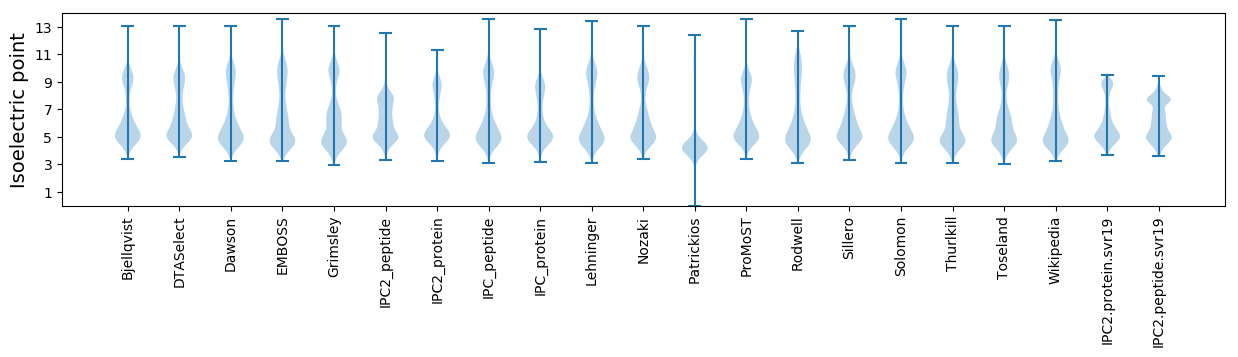
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A370DCS3|A0A370DCS3_9GAMM Uncharacterized protein OS=endosymbiont of Galathealinum brachiosum OX=2200906 GN=DIZ80_08780 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84IKK11 pKa = 10.5RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.96GFRR19 pKa = 11.84ARR21 pKa = 11.84SRR23 pKa = 11.84TVGGRR28 pKa = 11.84KK29 pKa = 8.24VLKK32 pKa = 10.25ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.31GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84IKK11 pKa = 10.5RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.96GFRR19 pKa = 11.84ARR21 pKa = 11.84SRR23 pKa = 11.84TVGGRR28 pKa = 11.84KK29 pKa = 8.24VLKK32 pKa = 10.25ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.31GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1105146 |
20 |
5022 |
323.7 |
36.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.482 ± 0.042 | 0.981 ± 0.015 |
6.094 ± 0.037 | 6.4 ± 0.049 |
4.087 ± 0.034 | 6.552 ± 0.049 |
2.167 ± 0.022 | 7.398 ± 0.035 |
6.161 ± 0.06 | 9.996 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.655 ± 0.025 | 5.13 ± 0.043 |
3.624 ± 0.032 | 3.938 ± 0.027 |
4.228 ± 0.033 | 7.03 ± 0.041 |
5.331 ± 0.046 | 6.569 ± 0.039 |
1.077 ± 0.015 | 3.101 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
