
Flammeovirga sp. SJP92
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Flammeovirgaceae; Flammeovirga; unclassified Flammeovirga
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
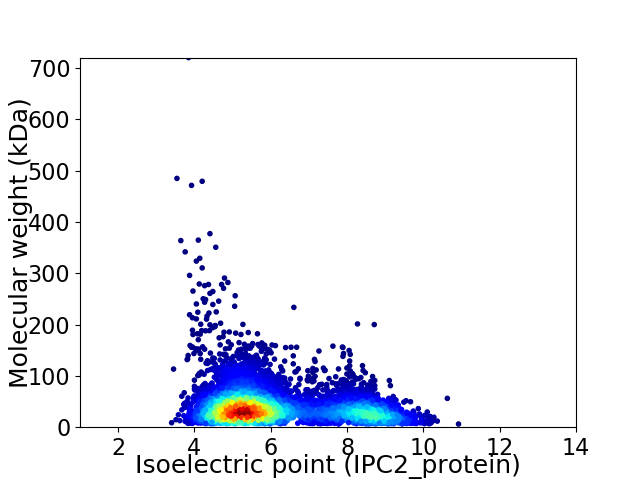
Virtual 2D-PAGE plot for 6269 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A150ARR8|A0A150ARR8_9BACT Uncharacterized protein OS=Flammeovirga sp. SJP92 OX=1775430 GN=AVL50_06405 PE=4 SV=1
MM1 pKa = 7.5LFATAVEE8 pKa = 4.24AQTTFRR14 pKa = 11.84PNKK17 pKa = 10.14LGTLNISSTGDD28 pKa = 2.82WQTASNWDD36 pKa = 3.64CGGCGSFPGSGDD48 pKa = 2.75IVLFGNDD55 pKa = 3.02NIYY58 pKa = 10.32WGDD61 pKa = 3.14AAIIIEE67 pKa = 4.26NVTLSGITFTASAEE81 pKa = 4.27GAALTIGSGATVNATSINEE100 pKa = 3.82IGKK103 pKa = 8.37NTISINSGGTLTINNSLTLDD123 pKa = 3.65NDD125 pKa = 3.53GTAFNVDD132 pKa = 2.74GDD134 pKa = 4.0VTINGDD140 pKa = 4.22LILSGGAAQTINSSGTLTVNGDD162 pKa = 3.45LSTEE166 pKa = 4.2GGSLNVTGGNVGITGDD182 pKa = 3.3WTIPSDD188 pKa = 4.13QANGNNGISISSGASVAIGGDD209 pKa = 3.11IAFLGNNSIPNPDD222 pKa = 3.22SQINIDD228 pKa = 4.16GSFTFNGSCTNNDD241 pKa = 3.82GGNDD245 pKa = 3.49LCQTFQDD252 pKa = 4.33INDD255 pKa = 3.96DD256 pKa = 3.87SLPVVLISLFGIQTEE271 pKa = 4.25EE272 pKa = 4.1GFLVKK277 pKa = 8.59WTTASEE283 pKa = 4.25QNNSHH288 pKa = 7.42FIIEE292 pKa = 4.71GSNSTDD298 pKa = 2.57NWKK301 pKa = 10.54EE302 pKa = 3.46IAEE305 pKa = 4.08VDD307 pKa = 3.79GQGNTNTSTDD317 pKa = 3.57YY318 pKa = 11.34EE319 pKa = 4.22IVLKK323 pKa = 10.87GEE325 pKa = 3.76QYY327 pKa = 10.78SYY329 pKa = 11.65YY330 pKa = 10.85RR331 pKa = 11.84LIQYY335 pKa = 10.33DD336 pKa = 3.57FDD338 pKa = 4.74GSFEE342 pKa = 3.99IFGILNQDD350 pKa = 2.73ISNTDD355 pKa = 3.05RR356 pKa = 11.84SVHH359 pKa = 5.7IFPNEE364 pKa = 3.69VNQGQFINLLFANGNADD381 pKa = 4.75LSITLQLYY389 pKa = 10.33DD390 pKa = 5.06LIGNVLDD397 pKa = 4.0YY398 pKa = 11.38QSVPNYY404 pKa = 10.3NSSLYY409 pKa = 9.72QYY411 pKa = 10.43KK412 pKa = 9.75VPNTAKK418 pKa = 10.77DD419 pKa = 3.1GGYY422 pKa = 10.55YY423 pKa = 9.76ILSVLIGNKK432 pKa = 9.51RR433 pKa = 11.84FHH435 pKa = 6.5QKK437 pKa = 10.29VLIKK441 pKa = 10.73
MM1 pKa = 7.5LFATAVEE8 pKa = 4.24AQTTFRR14 pKa = 11.84PNKK17 pKa = 10.14LGTLNISSTGDD28 pKa = 2.82WQTASNWDD36 pKa = 3.64CGGCGSFPGSGDD48 pKa = 2.75IVLFGNDD55 pKa = 3.02NIYY58 pKa = 10.32WGDD61 pKa = 3.14AAIIIEE67 pKa = 4.26NVTLSGITFTASAEE81 pKa = 4.27GAALTIGSGATVNATSINEE100 pKa = 3.82IGKK103 pKa = 8.37NTISINSGGTLTINNSLTLDD123 pKa = 3.65NDD125 pKa = 3.53GTAFNVDD132 pKa = 2.74GDD134 pKa = 4.0VTINGDD140 pKa = 4.22LILSGGAAQTINSSGTLTVNGDD162 pKa = 3.45LSTEE166 pKa = 4.2GGSLNVTGGNVGITGDD182 pKa = 3.3WTIPSDD188 pKa = 4.13QANGNNGISISSGASVAIGGDD209 pKa = 3.11IAFLGNNSIPNPDD222 pKa = 3.22SQINIDD228 pKa = 4.16GSFTFNGSCTNNDD241 pKa = 3.82GGNDD245 pKa = 3.49LCQTFQDD252 pKa = 4.33INDD255 pKa = 3.96DD256 pKa = 3.87SLPVVLISLFGIQTEE271 pKa = 4.25EE272 pKa = 4.1GFLVKK277 pKa = 8.59WTTASEE283 pKa = 4.25QNNSHH288 pKa = 7.42FIIEE292 pKa = 4.71GSNSTDD298 pKa = 2.57NWKK301 pKa = 10.54EE302 pKa = 3.46IAEE305 pKa = 4.08VDD307 pKa = 3.79GQGNTNTSTDD317 pKa = 3.57YY318 pKa = 11.34EE319 pKa = 4.22IVLKK323 pKa = 10.87GEE325 pKa = 3.76QYY327 pKa = 10.78SYY329 pKa = 11.65YY330 pKa = 10.85RR331 pKa = 11.84LIQYY335 pKa = 10.33DD336 pKa = 3.57FDD338 pKa = 4.74GSFEE342 pKa = 3.99IFGILNQDD350 pKa = 2.73ISNTDD355 pKa = 3.05RR356 pKa = 11.84SVHH359 pKa = 5.7IFPNEE364 pKa = 3.69VNQGQFINLLFANGNADD381 pKa = 4.75LSITLQLYY389 pKa = 10.33DD390 pKa = 5.06LIGNVLDD397 pKa = 4.0YY398 pKa = 11.38QSVPNYY404 pKa = 10.3NSSLYY409 pKa = 9.72QYY411 pKa = 10.43KK412 pKa = 9.75VPNTAKK418 pKa = 10.77DD419 pKa = 3.1GGYY422 pKa = 10.55YY423 pKa = 9.76ILSVLIGNKK432 pKa = 9.51RR433 pKa = 11.84FHH435 pKa = 6.5QKK437 pKa = 10.29VLIKK441 pKa = 10.73
Molecular weight: 46.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A150ADA6|A0A150ADA6_9BACT Uncharacterized protein OS=Flammeovirga sp. SJP92 OX=1775430 GN=AVL50_25135 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.89RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 10.04HH16 pKa = 4.44GFRR19 pKa = 11.84QRR21 pKa = 11.84MLTANGRR28 pKa = 11.84NVLAARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.6KK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.81SLTVSDD46 pKa = 4.03SRR48 pKa = 11.84KK49 pKa = 10.05HH50 pKa = 5.27KK51 pKa = 10.79AA52 pKa = 3.07
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.89RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 10.04HH16 pKa = 4.44GFRR19 pKa = 11.84QRR21 pKa = 11.84MLTANGRR28 pKa = 11.84NVLAARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.6KK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.81SLTVSDD46 pKa = 4.03SRR48 pKa = 11.84KK49 pKa = 10.05HH50 pKa = 5.27KK51 pKa = 10.79AA52 pKa = 3.07
Molecular weight: 6.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2376931 |
52 |
6570 |
379.2 |
42.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.755 ± 0.034 | 0.718 ± 0.011 |
5.826 ± 0.025 | 7.165 ± 0.029 |
5.122 ± 0.02 | 6.501 ± 0.031 |
1.908 ± 0.017 | 7.491 ± 0.031 |
7.514 ± 0.046 | 8.987 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.272 ± 0.015 | 5.977 ± 0.029 |
3.332 ± 0.019 | 3.65 ± 0.018 |
3.35 ± 0.018 | 6.8 ± 0.03 |
5.735 ± 0.041 | 6.262 ± 0.026 |
1.213 ± 0.011 | 4.422 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
