
Arhar cryptic virus-I
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
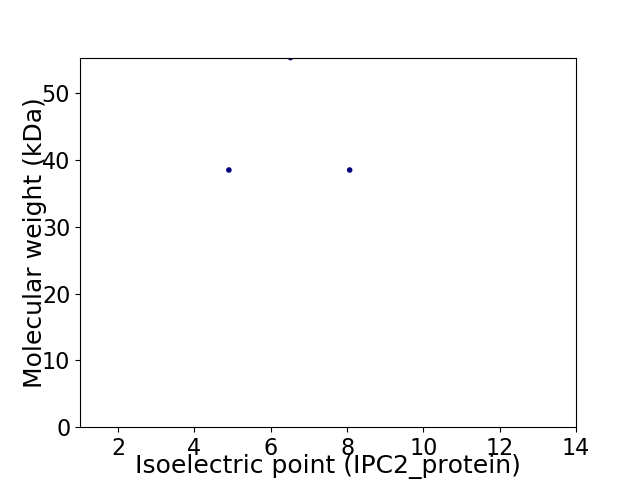
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|X5LY29|X5LY29_9VIRU Isoform of X5M5C8 Coat Protein OS=Arhar cryptic virus-I OX=1585924 GN=CP PE=4 SV=1
MM1 pKa = 7.31AAEE4 pKa = 5.26GPAQPSLTEE13 pKa = 4.42SGASSPNPPVVKK25 pKa = 9.82RR26 pKa = 11.84DD27 pKa = 3.52EE28 pKa = 5.39SKK30 pKa = 10.96DD31 pKa = 3.51QPKK34 pKa = 10.81ASIGSTFRR42 pKa = 11.84MKK44 pKa = 10.97NEE46 pKa = 3.63APVAQKK52 pKa = 10.88LVVALLDD59 pKa = 3.64QEE61 pKa = 4.8TVDD64 pKa = 4.76IPAAGNQNYY73 pKa = 9.95IIEE76 pKa = 4.4LKK78 pKa = 10.65EE79 pKa = 3.88KK80 pKa = 10.78GILQHH85 pKa = 6.81RR86 pKa = 11.84FPRR89 pKa = 11.84DD90 pKa = 3.28RR91 pKa = 11.84ISTVCEE97 pKa = 4.51PIPMNMQNLFTGALITIRR115 pKa = 11.84ASLTRR120 pKa = 11.84LLQLKK125 pKa = 7.37TQLGDD130 pKa = 3.13EE131 pKa = 4.64DD132 pKa = 5.0YY133 pKa = 8.17EE134 pKa = 4.32TRR136 pKa = 11.84LDD138 pKa = 3.78VVATNLAIGCNLVTFMKK155 pKa = 10.46LRR157 pKa = 11.84ALALTDD163 pKa = 3.21VEE165 pKa = 4.72TRR167 pKa = 11.84HH168 pKa = 5.98NADD171 pKa = 3.01GLRR174 pKa = 11.84RR175 pKa = 11.84PRR177 pKa = 11.84AIADD181 pKa = 3.56VPIPTAFAFAIQQLGIVNIADD202 pKa = 3.72LTHH205 pKa = 6.21EE206 pKa = 4.54LKK208 pKa = 10.55FVPCLPQPGYY218 pKa = 10.92QFGLPQTLTWNPNAYY233 pKa = 8.84QQAVEE238 pKa = 4.19YY239 pKa = 10.58ARR241 pKa = 11.84SLGMRR246 pKa = 11.84FSIVDD251 pKa = 3.5TTVKK255 pKa = 10.55EE256 pKa = 4.65GTAWWLFEE264 pKa = 3.61QSYY267 pKa = 10.36NEE269 pKa = 3.95NVFEE273 pKa = 4.75LMCPYY278 pKa = 10.23PEE280 pKa = 4.81SNFTEE285 pKa = 4.53MMALAHH291 pKa = 6.18SLYY294 pKa = 10.68LAGDD298 pKa = 3.54EE299 pKa = 4.71ADD301 pKa = 4.08PSNTLLNVEE310 pKa = 4.7DD311 pKa = 4.37MPHH314 pKa = 6.13GGNYY318 pKa = 9.76GIMMQFPHH326 pKa = 7.74LGIHH330 pKa = 6.43LSSFEE335 pKa = 4.26ALSEE339 pKa = 3.98AASDD343 pKa = 2.86IWSNVV348 pKa = 2.77
MM1 pKa = 7.31AAEE4 pKa = 5.26GPAQPSLTEE13 pKa = 4.42SGASSPNPPVVKK25 pKa = 9.82RR26 pKa = 11.84DD27 pKa = 3.52EE28 pKa = 5.39SKK30 pKa = 10.96DD31 pKa = 3.51QPKK34 pKa = 10.81ASIGSTFRR42 pKa = 11.84MKK44 pKa = 10.97NEE46 pKa = 3.63APVAQKK52 pKa = 10.88LVVALLDD59 pKa = 3.64QEE61 pKa = 4.8TVDD64 pKa = 4.76IPAAGNQNYY73 pKa = 9.95IIEE76 pKa = 4.4LKK78 pKa = 10.65EE79 pKa = 3.88KK80 pKa = 10.78GILQHH85 pKa = 6.81RR86 pKa = 11.84FPRR89 pKa = 11.84DD90 pKa = 3.28RR91 pKa = 11.84ISTVCEE97 pKa = 4.51PIPMNMQNLFTGALITIRR115 pKa = 11.84ASLTRR120 pKa = 11.84LLQLKK125 pKa = 7.37TQLGDD130 pKa = 3.13EE131 pKa = 4.64DD132 pKa = 5.0YY133 pKa = 8.17EE134 pKa = 4.32TRR136 pKa = 11.84LDD138 pKa = 3.78VVATNLAIGCNLVTFMKK155 pKa = 10.46LRR157 pKa = 11.84ALALTDD163 pKa = 3.21VEE165 pKa = 4.72TRR167 pKa = 11.84HH168 pKa = 5.98NADD171 pKa = 3.01GLRR174 pKa = 11.84RR175 pKa = 11.84PRR177 pKa = 11.84AIADD181 pKa = 3.56VPIPTAFAFAIQQLGIVNIADD202 pKa = 3.72LTHH205 pKa = 6.21EE206 pKa = 4.54LKK208 pKa = 10.55FVPCLPQPGYY218 pKa = 10.92QFGLPQTLTWNPNAYY233 pKa = 8.84QQAVEE238 pKa = 4.19YY239 pKa = 10.58ARR241 pKa = 11.84SLGMRR246 pKa = 11.84FSIVDD251 pKa = 3.5TTVKK255 pKa = 10.55EE256 pKa = 4.65GTAWWLFEE264 pKa = 3.61QSYY267 pKa = 10.36NEE269 pKa = 3.95NVFEE273 pKa = 4.75LMCPYY278 pKa = 10.23PEE280 pKa = 4.81SNFTEE285 pKa = 4.53MMALAHH291 pKa = 6.18SLYY294 pKa = 10.68LAGDD298 pKa = 3.54EE299 pKa = 4.71ADD301 pKa = 4.08PSNTLLNVEE310 pKa = 4.7DD311 pKa = 4.37MPHH314 pKa = 6.13GGNYY318 pKa = 9.76GIMMQFPHH326 pKa = 7.74LGIHH330 pKa = 6.43LSSFEE335 pKa = 4.26ALSEE339 pKa = 3.98AASDD343 pKa = 2.86IWSNVV348 pKa = 2.77
Molecular weight: 38.51 kDa
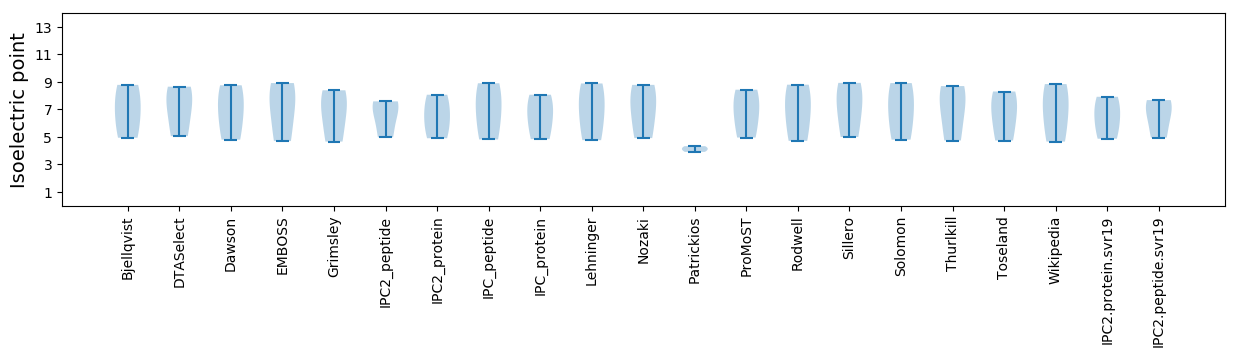
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X5LY29|X5LY29_9VIRU Isoform of X5M5C8 Coat Protein OS=Arhar cryptic virus-I OX=1585924 GN=CP PE=4 SV=1
MM1 pKa = 7.33VDD3 pKa = 3.03KK4 pKa = 10.97RR5 pKa = 11.84KK6 pKa = 8.82NTDD9 pKa = 3.06SQFASPTTAKK19 pKa = 9.37TPKK22 pKa = 9.56IVTPGEE28 pKa = 3.89PSNNNPALSIVTHH41 pKa = 6.75RR42 pKa = 11.84GASQAEE48 pKa = 4.58SIRR51 pKa = 11.84PEE53 pKa = 4.12AEE55 pKa = 3.96LYY57 pKa = 9.71PVQTKK62 pKa = 10.51RR63 pKa = 11.84LVLEE67 pKa = 4.79GNPSYY72 pKa = 10.71YY73 pKa = 10.23RR74 pKa = 11.84KK75 pKa = 10.27LLDD78 pKa = 3.77RR79 pKa = 11.84KK80 pKa = 9.5EE81 pKa = 3.52IGYY84 pKa = 9.97RR85 pKa = 11.84RR86 pKa = 11.84TGAGKK91 pKa = 10.1VLTFRR96 pKa = 11.84SFDD99 pKa = 3.89LNVSYY104 pKa = 10.54FRR106 pKa = 11.84TAITDD111 pKa = 3.18VMSRR115 pKa = 11.84VLKK118 pKa = 10.31IRR120 pKa = 11.84YY121 pKa = 8.43QSNGSMRR128 pKa = 11.84QTEE131 pKa = 4.43IDD133 pKa = 3.53TEE135 pKa = 3.99IATFVPHH142 pKa = 6.6IVNVCMSALYY152 pKa = 10.57AKK154 pKa = 10.21LRR156 pKa = 11.84SLNKK160 pKa = 9.77SYY162 pKa = 10.73GRR164 pKa = 11.84HH165 pKa = 4.0GTRR168 pKa = 11.84YY169 pKa = 8.13ATAPVYY175 pKa = 10.44TKK177 pKa = 10.31DD178 pKa = 3.58IEE180 pKa = 4.51IPLPLALAIQEE191 pKa = 4.37LGSFQTQNLEE201 pKa = 4.08NNVIMIPTYY210 pKa = 10.15PEE212 pKa = 4.53GVQHH216 pKa = 6.93EE217 pKa = 4.72GRR219 pKa = 11.84TAAEE223 pKa = 4.22FPVAEE228 pKa = 4.24YY229 pKa = 10.73LSYY232 pKa = 11.41VPTLLDD238 pKa = 3.7LKK240 pKa = 10.73IPLKK244 pKa = 10.99SVDD247 pKa = 3.32ARR249 pKa = 11.84IKK251 pKa = 9.88TGSAWWTYY259 pKa = 8.79KK260 pKa = 10.92LEE262 pKa = 4.62VIHH265 pKa = 5.55YY266 pKa = 6.81TADD269 pKa = 4.97FICTIPRR276 pKa = 11.84SLYY279 pKa = 10.8SDD281 pKa = 3.13MTALLRR287 pKa = 11.84TLFLSGSLEE296 pKa = 3.95QPPEE300 pKa = 4.03DD301 pKa = 3.24LVVFPEE307 pKa = 4.2DD308 pKa = 3.06QATFGTLLNCLSAGFNARR326 pKa = 11.84TFLALIGGPKK336 pKa = 9.85EE337 pKa = 3.78EE338 pKa = 4.55WSFGTNN344 pKa = 2.74
MM1 pKa = 7.33VDD3 pKa = 3.03KK4 pKa = 10.97RR5 pKa = 11.84KK6 pKa = 8.82NTDD9 pKa = 3.06SQFASPTTAKK19 pKa = 9.37TPKK22 pKa = 9.56IVTPGEE28 pKa = 3.89PSNNNPALSIVTHH41 pKa = 6.75RR42 pKa = 11.84GASQAEE48 pKa = 4.58SIRR51 pKa = 11.84PEE53 pKa = 4.12AEE55 pKa = 3.96LYY57 pKa = 9.71PVQTKK62 pKa = 10.51RR63 pKa = 11.84LVLEE67 pKa = 4.79GNPSYY72 pKa = 10.71YY73 pKa = 10.23RR74 pKa = 11.84KK75 pKa = 10.27LLDD78 pKa = 3.77RR79 pKa = 11.84KK80 pKa = 9.5EE81 pKa = 3.52IGYY84 pKa = 9.97RR85 pKa = 11.84RR86 pKa = 11.84TGAGKK91 pKa = 10.1VLTFRR96 pKa = 11.84SFDD99 pKa = 3.89LNVSYY104 pKa = 10.54FRR106 pKa = 11.84TAITDD111 pKa = 3.18VMSRR115 pKa = 11.84VLKK118 pKa = 10.31IRR120 pKa = 11.84YY121 pKa = 8.43QSNGSMRR128 pKa = 11.84QTEE131 pKa = 4.43IDD133 pKa = 3.53TEE135 pKa = 3.99IATFVPHH142 pKa = 6.6IVNVCMSALYY152 pKa = 10.57AKK154 pKa = 10.21LRR156 pKa = 11.84SLNKK160 pKa = 9.77SYY162 pKa = 10.73GRR164 pKa = 11.84HH165 pKa = 4.0GTRR168 pKa = 11.84YY169 pKa = 8.13ATAPVYY175 pKa = 10.44TKK177 pKa = 10.31DD178 pKa = 3.58IEE180 pKa = 4.51IPLPLALAIQEE191 pKa = 4.37LGSFQTQNLEE201 pKa = 4.08NNVIMIPTYY210 pKa = 10.15PEE212 pKa = 4.53GVQHH216 pKa = 6.93EE217 pKa = 4.72GRR219 pKa = 11.84TAAEE223 pKa = 4.22FPVAEE228 pKa = 4.24YY229 pKa = 10.73LSYY232 pKa = 11.41VPTLLDD238 pKa = 3.7LKK240 pKa = 10.73IPLKK244 pKa = 10.99SVDD247 pKa = 3.32ARR249 pKa = 11.84IKK251 pKa = 9.88TGSAWWTYY259 pKa = 8.79KK260 pKa = 10.92LEE262 pKa = 4.62VIHH265 pKa = 5.55YY266 pKa = 6.81TADD269 pKa = 4.97FICTIPRR276 pKa = 11.84SLYY279 pKa = 10.8SDD281 pKa = 3.13MTALLRR287 pKa = 11.84TLFLSGSLEE296 pKa = 3.95QPPEE300 pKa = 4.03DD301 pKa = 3.24LVVFPEE307 pKa = 4.2DD308 pKa = 3.06QATFGTLLNCLSAGFNARR326 pKa = 11.84TFLALIGGPKK336 pKa = 9.85EE337 pKa = 3.78EE338 pKa = 4.55WSFGTNN344 pKa = 2.74
Molecular weight: 38.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1167 |
344 |
475 |
389.0 |
44.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.055 ± 0.628 | 0.943 ± 0.075 |
4.799 ± 0.403 | 6.512 ± 0.151 |
4.37 ± 0.234 | 4.97 ± 0.377 |
2.485 ± 0.555 | 6.17 ± 0.392 |
4.113 ± 0.362 | 10.026 ± 0.288 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.399 ± 0.373 | 4.37 ± 0.45 |
5.57 ± 0.785 | 3.599 ± 0.583 |
6.255 ± 0.841 | 7.112 ± 0.535 |
6.341 ± 1.099 | 5.998 ± 0.089 |
1.457 ± 0.327 | 4.456 ± 0.736 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
